How To Write Significance of the Study (With Examples)

Whether you’re writing a research paper or thesis, a portion called Significance of the Study ensures your readers understand the impact of your work. Learn how to effectively write this vital part of your research paper or thesis through our detailed steps, guidelines, and examples.
Related: How to Write a Concept Paper for Academic Research

Table of Contents
What is the significance of the study.
The Significance of the Study presents the importance of your research. It allows you to prove the study’s impact on your field of research, the new knowledge it contributes, and the people who will benefit from it.
Related: How To Write Scope and Delimitation of a Research Paper (With Examples)
Where Should I Put the Significance of the Study?
The Significance of the Study is part of the first chapter or the Introduction. It comes after the research’s rationale, problem statement, and hypothesis.
Related: How to Make Conceptual Framework (with Examples and Templates)
Why Should I Include the Significance of the Study?
The purpose of the Significance of the Study is to give you space to explain to your readers how exactly your research will be contributing to the literature of the field you are studying 1 . It’s where you explain why your research is worth conducting and its significance to the community, the people, and various institutions.
How To Write Significance of the Study: 5 Steps
Below are the steps and guidelines for writing your research’s Significance of the Study.
1. Use Your Research Problem as a Starting Point
Your problem statement can provide clues to your research study’s outcome and who will benefit from it 2 .
Ask yourself, “How will the answers to my research problem be beneficial?”. In this manner, you will know how valuable it is to conduct your study.
Let’s say your research problem is “What is the level of effectiveness of the lemongrass (Cymbopogon citratus) in lowering the blood glucose level of Swiss mice (Mus musculus)?”
Discovering a positive correlation between the use of lemongrass and lower blood glucose level may lead to the following results:
- Increased public understanding of the plant’s medical properties;
- Higher appreciation of the importance of lemongrass by the community;
- Adoption of lemongrass tea as a cheap, readily available, and natural remedy to lower their blood glucose level.
Once you’ve zeroed in on the general benefits of your study, it’s time to break it down into specific beneficiaries.
2. State How Your Research Will Contribute to the Existing Literature in the Field
Think of the things that were not explored by previous studies. Then, write how your research tackles those unexplored areas. Through this, you can convince your readers that you are studying something new and adding value to the field.
3. Explain How Your Research Will Benefit Society
In this part, tell how your research will impact society. Think of how the results of your study will change something in your community.
For example, in the study about using lemongrass tea to lower blood glucose levels, you may indicate that through your research, the community will realize the significance of lemongrass and other herbal plants. As a result, the community will be encouraged to promote the cultivation and use of medicinal plants.
4. Mention the Specific Persons or Institutions Who Will Benefit From Your Study
Using the same example above, you may indicate that this research’s results will benefit those seeking an alternative supplement to prevent high blood glucose levels.
5. Indicate How Your Study May Help Future Studies in the Field
You must also specifically indicate how your research will be part of the literature of your field and how it will benefit future researchers. In our example above, you may indicate that through the data and analysis your research will provide, future researchers may explore other capabilities of herbal plants in preventing different diseases.
Tips and Warnings
- Think ahead . By visualizing your study in its complete form, it will be easier for you to connect the dots and identify the beneficiaries of your research.
- Write concisely. Make it straightforward, clear, and easy to understand so that the readers will appreciate the benefits of your research. Avoid making it too long and wordy.
- Go from general to specific . Like an inverted pyramid, you start from above by discussing the general contribution of your study and become more specific as you go along. For instance, if your research is about the effect of remote learning setup on the mental health of college students of a specific university , you may start by discussing the benefits of the research to society, to the educational institution, to the learning facilitators, and finally, to the students.
- Seek help . For example, you may ask your research adviser for insights on how your research may contribute to the existing literature. If you ask the right questions, your research adviser can point you in the right direction.
- Revise, revise, revise. Be ready to apply necessary changes to your research on the fly. Unexpected things require adaptability, whether it’s the respondents or variables involved in your study. There’s always room for improvement, so never assume your work is done until you have reached the finish line.
Significance of the Study Examples
This section presents examples of the Significance of the Study using the steps and guidelines presented above.
Example 1: STEM-Related Research
Research Topic: Level of Effectiveness of the Lemongrass ( Cymbopogon citratus ) Tea in Lowering the Blood Glucose Level of Swiss Mice ( Mus musculus ).
Significance of the Study .
This research will provide new insights into the medicinal benefit of lemongrass ( Cymbopogon citratus ), specifically on its hypoglycemic ability.
Through this research, the community will further realize promoting medicinal plants, especially lemongrass, as a preventive measure against various diseases. People and medical institutions may also consider lemongrass tea as an alternative supplement against hyperglycemia.
Moreover, the analysis presented in this study will convey valuable information for future research exploring the medicinal benefits of lemongrass and other medicinal plants.
Example 2: Business and Management-Related Research
Research Topic: A Comparative Analysis of Traditional and Social Media Marketing of Small Clothing Enterprises.
Significance of the Study:
By comparing the two marketing strategies presented by this research, there will be an expansion on the current understanding of the firms on these marketing strategies in terms of cost, acceptability, and sustainability. This study presents these marketing strategies for small clothing enterprises, giving them insights into which method is more appropriate and valuable for them.
Specifically, this research will benefit start-up clothing enterprises in deciding which marketing strategy they should employ. Long-time clothing enterprises may also consider the result of this research to review their current marketing strategy.
Furthermore, a detailed presentation on the comparison of the marketing strategies involved in this research may serve as a tool for further studies to innovate the current method employed in the clothing Industry.
Example 3: Social Science -Related Research.
Research Topic: Divide Et Impera : An Overview of How the Divide-and-Conquer Strategy Prevailed on Philippine Political History.
Significance of the Study :
Through the comprehensive exploration of this study on Philippine political history, the influence of the Divide et Impera, or political decentralization, on the political discernment across the history of the Philippines will be unraveled, emphasized, and scrutinized. Moreover, this research will elucidate how this principle prevailed until the current political theatre of the Philippines.
In this regard, this study will give awareness to society on how this principle might affect the current political context. Moreover, through the analysis made by this study, political entities and institutions will have a new approach to how to deal with this principle by learning about its influence in the past.
In addition, the overview presented in this research will push for new paradigms, which will be helpful for future discussion of the Divide et Impera principle and may lead to a more in-depth analysis.
Example 4: Humanities-Related Research
Research Topic: Effectiveness of Meditation on Reducing the Anxiety Levels of College Students.
Significance of the Study:
This research will provide new perspectives in approaching anxiety issues of college students through meditation.
Specifically, this research will benefit the following:
Community – this study spreads awareness on recognizing anxiety as a mental health concern and how meditation can be a valuable approach to alleviating it.
Academic Institutions and Administrators – through this research, educational institutions and administrators may promote programs and advocacies regarding meditation to help students deal with their anxiety issues.
Mental health advocates – the result of this research will provide valuable information for the advocates to further their campaign on spreading awareness on dealing with various mental health issues, including anxiety, and how to stop stigmatizing those with mental health disorders.
Parents – this research may convince parents to consider programs involving meditation that may help the students deal with their anxiety issues.
Students will benefit directly from this research as its findings may encourage them to consider meditation to lower anxiety levels.
Future researchers – this study covers information involving meditation as an approach to reducing anxiety levels. Thus, the result of this study can be used for future discussions on the capabilities of meditation in alleviating other mental health concerns.
Frequently Asked Questions
1. what is the difference between the significance of the study and the rationale of the study.
Both aim to justify the conduct of the research. However, the Significance of the Study focuses on the specific benefits of your research in the field, society, and various people and institutions. On the other hand, the Rationale of the Study gives context on why the researcher initiated the conduct of the study.
Let’s take the research about the Effectiveness of Meditation in Reducing Anxiety Levels of College Students as an example. Suppose you are writing about the Significance of the Study. In that case, you must explain how your research will help society, the academic institution, and students deal with anxiety issues through meditation. Meanwhile, for the Rationale of the Study, you may state that due to the prevalence of anxiety attacks among college students, you’ve decided to make it the focal point of your research work.
2. What is the difference between Justification and the Significance of the Study?
In Justification, you express the logical reasoning behind the conduct of the study. On the other hand, the Significance of the Study aims to present to your readers the specific benefits your research will contribute to the field you are studying, community, people, and institutions.
Suppose again that your research is about the Effectiveness of Meditation in Reducing the Anxiety Levels of College Students. Suppose you are writing the Significance of the Study. In that case, you may state that your research will provide new insights and evidence regarding meditation’s ability to reduce college students’ anxiety levels. Meanwhile, you may note in the Justification that studies are saying how people used meditation in dealing with their mental health concerns. You may also indicate how meditation is a feasible approach to managing anxiety using the analysis presented by previous literature.
3. How should I start my research’s Significance of the Study section?
– This research will contribute… – The findings of this research… – This study aims to… – This study will provide… – Through the analysis presented in this study… – This study will benefit…
Moreover, you may start the Significance of the Study by elaborating on the contribution of your research in the field you are studying.
4. What is the difference between the Purpose of the Study and the Significance of the Study?
The Purpose of the Study focuses on why your research was conducted, while the Significance of the Study tells how the results of your research will benefit anyone.
Suppose your research is about the Effectiveness of Lemongrass Tea in Lowering the Blood Glucose Level of Swiss Mice . You may include in your Significance of the Study that the research results will provide new information and analysis on the medical ability of lemongrass to solve hyperglycemia. Meanwhile, you may include in your Purpose of the Study that your research wants to provide a cheaper and natural way to lower blood glucose levels since commercial supplements are expensive.
5. What is the Significance of the Study in Tagalog?
In Filipino research, the Significance of the Study is referred to as Kahalagahan ng Pag-aaral.
- Draft your Significance of the Study. Retrieved 18 April 2021, from http://dissertationedd.usc.edu/draft-your-significance-of-the-study.html
- Regoniel, P. (2015). Two Tips on How to Write the Significance of the Study. Retrieved 18 April 2021, from https://simplyeducate.me/2015/02/09/significance-of-the-study/
Written by Jewel Kyle Fabula
in Career and Education , Juander How
Jewel Kyle Fabula
Jewel Kyle Fabula is a Bachelor of Science in Economics student at the University of the Philippines Diliman. His passion for learning mathematics developed as he competed in some mathematics competitions during his Junior High School years. He loves cats, playing video games, and listening to music.
Browse all articles written by Jewel Kyle Fabula
Copyright Notice
All materials contained on this site are protected by the Republic of the Philippines copyright law and may not be reproduced, distributed, transmitted, displayed, published, or broadcast without the prior written permission of filipiknow.net or in the case of third party materials, the owner of that content. You may not alter or remove any trademark, copyright, or other notice from copies of the content. Be warned that we have already reported and helped terminate several websites and YouTube channels for blatantly stealing our content. If you wish to use filipiknow.net content for commercial purposes, such as for content syndication, etc., please contact us at legal(at)filipiknow(dot)net

The Savvy Scientist
Experiences of a London PhD student and beyond
What is the Significance of a Study? Examples and Guide

If you’re reading this post you’re probably wondering: what is the significance of a study?
No matter where you’re at with a piece of research, it is a good idea to think about the potential significance of your work. And sometimes you’ll have to explicitly write a statement of significance in your papers, it addition to it forming part of your thesis.
In this post I’ll cover what the significance of a study is, how to measure it, how to describe it with examples and add in some of my own experiences having now worked in research for over nine years.
If you’re reading this because you’re writing up your first paper, welcome! You may also like my how-to guide for all aspects of writing your first research paper .
Looking for guidance on writing the statement of significance for a paper or thesis? Click here to skip straight to that section.
What is the Significance of a Study?
For research papers, theses or dissertations it’s common to explicitly write a section describing the significance of the study. We’ll come onto what to include in that section in just a moment.
However the significance of a study can actually refer to several different things.

Working our way from the most technical to the broadest, depending on the context, the significance of a study may refer to:
- Within your study: Statistical significance. Can we trust the findings?
- Wider research field: Research significance. How does your study progress the field?
- Commercial / economic significance: Could there be business opportunities for your findings?
- Societal significance: What impact could your study have on the wider society.
- And probably other domain-specific significance!
We’ll shortly cover each of them in turn, including how they’re measured and some examples for each type of study significance.
But first, let’s touch on why you should consider the significance of your research at an early stage.
Why Care About the Significance of a Study?
No matter what is motivating you to carry out your research, it is sensible to think about the potential significance of your work. In the broadest sense this asks, how does the study contribute to the world?
After all, for many people research is only worth doing if it will result in some expected significance. For the vast majority of us our studies won’t be significant enough to reach the evening news, but most studies will help to enhance knowledge in a particular field and when research has at least some significance it makes for a far more fulfilling longterm pursuit.
Furthermore, a lot of us are carrying out research funded by the public. It therefore makes sense to keep an eye on what benefits the work could bring to the wider community.
Often in research you’ll come to a crossroads where you must decide which path of research to pursue. Thinking about the potential benefits of a strand of research can be useful for deciding how to spend your time, money and resources.
It’s worth noting though, that not all research activities have to work towards obvious significance. This is especially true while you’re a PhD student, where you’re figuring out what you enjoy and may simply be looking for an opportunity to learn a new skill.
However, if you’re trying to decide between two potential projects, it can be useful to weigh up the potential significance of each.
Let’s now dive into the different types of significance, starting with research significance.
Research Significance
What is the research significance of a study.
Unless someone specifies which type of significance they’re referring to, it is fair to assume that they want to know about the research significance of your study.
Research significance describes how your work has contributed to the field, how it could inform future studies and progress research.
Where should I write about my study’s significance in my thesis?
Typically you should write about your study’s significance in the Introduction and Conclusions sections of your thesis.
It’s important to mention it in the Introduction so that the relevance of your work and the potential impact and benefits it could have on the field are immediately apparent. Explaining why your work matters will help to engage readers (and examiners!) early on.
It’s also a good idea to detail the study’s significance in your Conclusions section. This adds weight to your findings and helps explain what your study contributes to the field.
On occasion you may also choose to include a brief description in your Abstract.
What is expected when submitting an article to a journal
It is common for journals to request a statement of significance, although this can sometimes be called other things such as:
- Impact statement
- Significance statement
- Advances in knowledge section
Here is one such example of what is expected:
Impact Statement: An Impact Statement is required for all submissions. Your impact statement will be evaluated by the Editor-in-Chief, Global Editors, and appropriate Associate Editor. For your manuscript to receive full review, the editors must be convinced that it is an important advance in for the field. The Impact Statement is not a restating of the abstract. It should address the following: Why is the work submitted important to the field? How does the work submitted advance the field? What new information does this work impart to the field? How does this new information impact the field? Experimental Biology and Medicine journal, author guidelines
Typically the impact statement will be shorter than the Abstract, around 150 words.
Defining the study’s significance is helpful not just for the impact statement (if the journal asks for one) but also for building a more compelling argument throughout your submission. For instance, usually you’ll start the Discussion section of a paper by highlighting the research significance of your work. You’ll also include a short description in your Abstract too.
How to describe the research significance of a study, with examples
Whether you’re writing a thesis or a journal article, the approach to writing about the significance of a study are broadly the same.
I’d therefore suggest using the questions above as a starting point to base your statements on.
- Why is the work submitted important to the field?
- How does the work submitted advance the field?
- What new information does this work impart to the field?
- How does this new information impact the field?
Answer those questions and you’ll have a much clearer idea of the research significance of your work.
When describing it, try to clearly state what is novel about your study’s contribution to the literature. Then go on to discuss what impact it could have on progressing the field along with recommendations for future work.
Potential sentence starters
If you’re not sure where to start, why not set a 10 minute timer and have a go at trying to finish a few of the following sentences. Not sure on what to put? Have a chat to your supervisor or lab mates and they may be able to suggest some ideas.
- This study is important to the field because…
- These findings advance the field by…
- Our results highlight the importance of…
- Our discoveries impact the field by…
Now you’ve had a go let’s have a look at some real life examples.
Statement of significance examples
A statement of significance / impact:
Impact Statement This review highlights the historical development of the concept of “ideal protein” that began in the 1950s and 1980s for poultry and swine diets, respectively, and the major conceptual deficiencies of the long-standing concept of “ideal protein” in animal nutrition based on recent advances in amino acid (AA) metabolism and functions. Nutritionists should move beyond the “ideal protein” concept to consider optimum ratios and amounts of all proteinogenic AAs in animal foods and, in the case of carnivores, also taurine. This will help formulate effective low-protein diets for livestock, poultry, and fish, while sustaining global animal production. Because they are not only species of agricultural importance, but also useful models to study the biology and diseases of humans as well as companion (e.g. dogs and cats), zoo, and extinct animals in the world, our work applies to a more general readership than the nutritionists and producers of farm animals. Wu G, Li P. The “ideal protein” concept is not ideal in animal nutrition. Experimental Biology and Medicine . 2022;247(13):1191-1201. doi: 10.1177/15353702221082658
And the same type of section but this time called “Advances in knowledge”:
Advances in knowledge: According to the MY-RADs criteria, size measurements of focal lesions in MRI are now of relevance for response assessment in patients with monoclonal plasma cell disorders. Size changes of 1 or 2 mm are frequently observed due to uncertainty of the measurement only, while the actual focal lesion has not undergone any biological change. Size changes of at least 6 mm or more in T 1 weighted or T 2 weighted short tau inversion recovery sequences occur in only 5% or less of cases when the focal lesion has not undergone any biological change. Wennmann M, Grözinger M, Weru V, et al. Test-retest, inter- and intra-rater reproducibility of size measurements of focal bone marrow lesions in MRI in patients with multiple myeloma [published online ahead of print, 2023 Apr 12]. Br J Radiol . 2023;20220745. doi: 10.1259/bjr.20220745
Other examples of research significance
Moving beyond the formal statement of significance, here is how you can describe research significance more broadly within your paper.
Describing research impact in an Abstract of a paper:
Three-dimensional visualisation and quantification of the chondrocyte population within articular cartilage can be achieved across a field of view of several millimetres using laboratory-based micro-CT. The ability to map chondrocytes in 3D opens possibilities for research in fields from skeletal development through to medical device design and treatment of cartilage degeneration. Conclusions section of the abstract in my first paper .
In the Discussion section of a paper:
We report for the utility of a standard laboratory micro-CT scanner to visualise and quantify features of the chondrocyte population within intact articular cartilage in 3D. This study represents a complimentary addition to the growing body of evidence supporting the non-destructive imaging of the constituents of articular cartilage. This offers researchers the opportunity to image chondrocyte distributions in 3D without specialised synchrotron equipment, enabling investigations such as chondrocyte morphology across grades of cartilage damage, 3D strain mapping techniques such as digital volume correlation to evaluate mechanical properties in situ , and models for 3D finite element analysis in silico simulations. This enables an objective quantification of chondrocyte distribution and morphology in three dimensions allowing greater insight for investigations into studies of cartilage development, degeneration and repair. One such application of our method, is as a means to provide a 3D pattern in the cartilage which, when combined with digital volume correlation, could determine 3D strain gradient measurements enabling potential treatment and repair of cartilage degeneration. Moreover, the method proposed here will allow evaluation of cartilage implanted with tissue engineered scaffolds designed to promote chondral repair, providing valuable insight into the induced regenerative process. The Discussion section of the paper is laced with references to research significance.
How is longer term research significance measured?
Looking beyond writing impact statements within papers, sometimes you’ll want to quantify the long term research significance of your work. For instance when applying for jobs.
The most obvious measure of a study’s long term research significance is the number of citations it receives from future publications. The thinking is that a study which receives more citations will have had more research impact, and therefore significance , than a study which received less citations. Citations can give a broad indication of how useful the work is to other researchers but citations aren’t really a good measure of significance.
Bear in mind that us researchers can be lazy folks and sometimes are simply looking to cite the first paper which backs up one of our claims. You can find studies which receive a lot of citations simply for packaging up the obvious in a form which can be easily found and referenced, for instance by having a catchy or optimised title.
Likewise, research activity varies wildly between fields. Therefore a certain study may have had a big impact on a particular field but receive a modest number of citations, simply because not many other researchers are working in the field.
Nevertheless, citations are a standard measure of significance and for better or worse it remains impressive for someone to be the first author of a publication receiving lots of citations.
Other measures for the research significance of a study include:
- Accolades: best paper awards at conferences, thesis awards, “most downloaded” titles for articles, press coverage.
- How much follow-on research the study creates. For instance, part of my PhD involved a novel material initially developed by another PhD student in the lab. That PhD student’s research had unlocked lots of potential new studies and now lots of people in the group were using the same material and developing it for different applications. The initial study may not receive a high number of citations yet long term it generated a lot of research activity.
That covers research significance, but you’ll often want to consider other types of significance for your study and we’ll cover those next.
Statistical Significance
What is the statistical significance of a study.
Often as part of a study you’ll carry out statistical tests and then state the statistical significance of your findings: think p-values eg <0.05. It is useful to describe the outcome of these tests within your report or paper, to give a measure of statistical significance.
Effectively you are trying to show whether the performance of your innovation is actually better than a control or baseline and not just chance. Statistical significance deserves a whole other post so I won’t go into a huge amount of depth here.
Things that make publication in The BMJ impossible or unlikely Internal validity/robustness of the study • It had insufficient statistical power, making interpretation difficult; • Lack of statistical power; The British Medical Journal’s guide for authors
Calculating statistical significance isn’t always necessary (or valid) for a study, such as if you have a very small number of samples, but it is a very common requirement for scientific articles.
Writing a journal article? Check the journal’s guide for authors to see what they expect. Generally if you have approximately five or more samples or replicates it makes sense to start thinking about statistical tests. Speak to your supervisor and lab mates for advice, and look at other published articles in your field.
How is statistical significance measured?
Statistical significance is quantified using p-values . Depending on your study design you’ll choose different statistical tests to compute the p-value.
A p-value of 0.05 is a common threshold value. The 0.05 means that there is a 1/20 chance that the difference in performance you’re reporting is just down to random chance.
- p-values above 0.05 mean that the result isn’t statistically significant enough to be trusted: it is too likely that the effect you’re showing is just luck.
- p-values less than or equal to 0.05 mean that the result is statistically significant. In other words: unlikely to just be chance, which is usually considered a good outcome.
Low p-values (eg p = 0.001) mean that it is highly unlikely to be random chance (1/1000 in the case of p = 0.001), therefore more statistically significant.
It is important to clarify that, although low p-values mean that your findings are statistically significant, it doesn’t automatically mean that the result is scientifically important. More on that in the next section on research significance.
How to describe the statistical significance of your study, with examples
In the first paper from my PhD I ran some statistical tests to see if different staining techniques (basically dyes) increased how well you could see cells in cow tissue using micro-CT scanning (a 3D imaging technique).
In your methods section you should mention the statistical tests you conducted and then in the results you will have statements such as:
Between mediums for the two scan protocols C/N [contrast to noise ratio] was greater for EtOH than the PBS in both scanning methods (both p < 0.0001) with mean differences of 1.243 (95% CI [confidence interval] 0.709 to 1.778) for absorption contrast and 6.231 (95% CI 5.772 to 6.690) for propagation contrast. … Two repeat propagation scans were taken of samples from the PTA-stained groups. No difference in mean C/N was found with either medium: PBS had a mean difference of 0.058 ( p = 0.852, 95% CI -0.560 to 0.676), EtOH had a mean difference of 1.183 ( p = 0.112, 95% CI 0.281 to 2.648). From the Results section of my first paper, available here . Square brackets added for this post to aid clarity.
From this text the reader can infer from the first paragraph that there was a statistically significant difference in using EtOH compared to PBS (really small p-value of <0.0001). However, from the second paragraph, the difference between two repeat scans was statistically insignificant for both PBS (p = 0.852) and EtOH (p = 0.112).
By conducting these statistical tests you have then earned your right to make bold statements, such as these from the discussion section:
Propagation phase-contrast increases the contrast of individual chondrocytes [cartilage cells] compared to using absorption contrast. From the Discussion section from the same paper.
Without statistical tests you have no evidence that your results are not just down to random chance.
Beyond describing the statistical significance of a study in the main body text of your work, you can also show it in your figures.
In figures such as bar charts you’ll often see asterisks to represent statistical significance, and “n.s.” to show differences between groups which are not statistically significant. Here is one such figure, with some subplots, from the same paper:

In this example an asterisk (*) between two bars represents p < 0.05. Two asterisks (**) represents p < 0.001 and three asterisks (***) represents p < 0.0001. This should always be stated in the caption of your figure since the values that each asterisk refers to can vary.
Now that we know if a study is showing statistically and research significance, let’s zoom out a little and consider the potential for commercial significance.
Commercial and Industrial Significance
What are commercial and industrial significance.
Moving beyond significance in relation to academia, your research may also have commercial or economic significance.
Simply put:
- Commercial significance: could the research be commercialised as a product or service? Perhaps the underlying technology described in your study could be licensed to a company or you could even start your own business using it.
- Industrial significance: more widely than just providing a product which could be sold, does your research provide insights which may affect a whole industry? Such as: revealing insights or issues with current practices, performance gains you don’t want to commercialise (e.g. solar power efficiency), providing suggested frameworks or improvements which could be employed industry-wide.
I’ve grouped these two together because there can certainly be overlap. For instance, perhaps your new technology could be commercialised whilst providing wider improvements for the whole industry.
Commercial and industrial significance are not relevant to most studies, so only write about it if you and your supervisor can think of reasonable routes to your work having an impact in these ways.
How are commercial and industrial significance measured?
Unlike statistical and research significances, the measures of commercial and industrial significance can be much more broad.
Here are some potential measures of significance:
Commercial significance:
- How much value does your technology bring to potential customers or users?
- How big is the potential market and how much revenue could the product potentially generate?
- Is the intellectual property protectable? i.e. patentable, or if not could the novelty be protected with trade secrets: if so publish your method with caution!
- If commercialised, could the product bring employment to a geographical area?
Industrial significance:
What impact could it have on the industry? For instance if you’re revealing an issue with something, such as unintended negative consequences of a drug , what does that mean for the industry and the public? This could be:
- Reduced overhead costs
- Better safety
- Faster production methods
- Improved scaleability
How to describe the commercial and industrial significance of a study, with examples
Commercial significance.
If your technology could be commercially viable, and you’ve got an interest in commercialising it yourself, it is likely that you and your university may not want to immediately publish the study in a journal.
You’ll probably want to consider routes to exploiting the technology and your university may have a “technology transfer” team to help researchers navigate the various options.
However, if instead of publishing a paper you’re submitting a thesis or dissertation then it can be useful to highlight the commercial significance of your work. In this instance you could include statements of commercial significance such as:
The measurement technology described in this study provides state of the art performance and could enable the development of low cost devices for aerospace applications. An example of commercial significance I invented for this post
Industrial significance
First, think about the industrial sectors who could benefit from the developments described in your study.
For example if you’re working to improve battery efficiency it is easy to think of how it could lead to performance gains for certain industries, like personal electronics or electric vehicles. In these instances you can describe the industrial significance relatively easily, based off your findings.
For example:
By utilising abundant materials in the described battery fabrication process we provide a framework for battery manufacturers to reduce dependence on rare earth components. Again, an invented example
For other technologies there may well be industrial applications but they are less immediately obvious and applicable. In these scenarios the best you can do is to simply reframe your research significance statement in terms of potential commercial applications in a broad way.
As a reminder: not all studies should address industrial significance, so don’t try to invent applications just for the sake of it!
Societal Significance
What is the societal significance of a study.
The most broad category of significance is the societal impact which could stem from it.
If you’re working in an applied field it may be quite easy to see a route for your research to impact society. For others, the route to societal significance may be less immediate or clear.
Studies can help with big issues facing society such as:
- Medical applications : vaccines, surgical implants, drugs, improving patient safety. For instance this medical device and drug combination I worked on which has a very direct route to societal significance.
- Political significance : Your research may provide insights which could contribute towards potential changes in policy or better understanding of issues facing society.
- Public health : for instance COVID-19 transmission and related decisions.
- Climate change : mitigation such as more efficient solar panels and lower cost battery solutions, and studying required adaptation efforts and technologies. Also, better understanding around related societal issues, for instance this study on the effects of temperature on hate speech.
How is societal significance measured?
Societal significance at a high level can be quantified by the size of its potential societal effect. Just like a lab risk assessment, you can think of it in terms of probability (or how many people it could help) and impact magnitude.
Societal impact = How many people it could help x the magnitude of the impact
Think about how widely applicable the findings are: for instance does it affect only certain people? Then think about the potential size of the impact: what kind of difference could it make to those people?
Between these two metrics you can get a pretty good overview of the potential societal significance of your research study.
How to describe the societal significance of a study, with examples
Quite often the broad societal significance of your study is what you’re setting the scene for in your Introduction. In addition to describing the existing literature, it is common to for the study’s motivation to touch on its wider impact for society.
For those of us working in healthcare research it is usually pretty easy to see a path towards societal significance.
Our CLOUT model has state-of-the-art performance in mortality prediction, surpassing other competitive NN models and a logistic regression model … Our results show that the risk factors identified by the CLOUT model agree with physicians’ assessment, suggesting that CLOUT could be used in real-world clinicalsettings. Our results strongly support that CLOUT may be a useful tool to generate clinical prediction models, especially among hospitalized and critically ill patient populations. Learning Latent Space Representations to Predict Patient Outcomes: Model Development and Validation
In other domains the societal significance may either take longer or be more indirect, meaning that it can be more difficult to describe the societal impact.
Even so, here are some examples I’ve found from studies in non-healthcare domains:
We examined food waste as an initial investigation and test of this methodology, and there is clear potential for the examination of not only other policy texts related to food waste (e.g., liability protection, tax incentives, etc.; Broad Leib et al., 2020) but related to sustainable fishing (Worm et al., 2006) and energy use (Hawken, 2017). These other areas are of obvious relevance to climate change… AI-Based Text Analysis for Evaluating Food Waste Policies
The continued development of state-of-the art NLP tools tailored to climate policy will allow climate researchers and policy makers to extract meaningful information from this growing body of text, to monitor trends over time and administrative units, and to identify potential policy improvements. BERT Classification of Paris Agreement Climate Action Plans
Top Tips For Identifying & Writing About the Significance of Your Study
- Writing a thesis? Describe the significance of your study in the Introduction and the Conclusion .
- Submitting a paper? Read the journal’s guidelines. If you’re writing a statement of significance for a journal, make sure you read any guidance they give for what they’re expecting.
- Take a step back from your research and consider your study’s main contributions.
- Read previously published studies in your field . Use this for inspiration and ideas on how to describe the significance of your own study
- Discuss the study with your supervisor and potential co-authors or collaborators and brainstorm potential types of significance for it.
Now you’ve finished reading up on the significance of a study you may also like my how-to guide for all aspects of writing your first research paper .
Writing an academic journal paper
I hope that you’ve learned something useful from this article about the significance of a study. If you have any more research-related questions let me know, I’m here to help.
To gain access to my content library you can subscribe below for free:
Share this:
- Click to share on Facebook (Opens in new window)
- Click to share on LinkedIn (Opens in new window)
- Click to share on Twitter (Opens in new window)
- Click to share on Reddit (Opens in new window)
Related Posts

How to Plan a Research Visit
13th September 2024 13th September 2024

The Five Most Powerful Lessons I Learned During My PhD
8th August 2024 8th August 2024

Minor Corrections: How To Make Them and Succeed With Your PhD Thesis
2nd June 2024 2nd June 2024
Leave a Reply Cancel reply
Your email address will not be published. Required fields are marked *
Notify me of follow-up comments by email.
This site uses Akismet to reduce spam. Learn how your comment data is processed .
Privacy Overview

Significance of the Study
Ai generator.
The significance of the study underscores the research’s importance, illustrating its impact on existing knowledge and potential applications. It highlights how the findings address gaps, resolve problems, or contribute to advancements in a specific field. By emphasizing the study’s relevance, it demonstrates the broader implications for society, academia, or industry, justifying the research effort and investment.
What is the Significance of the Study?
The significance of the study illustrates the research’s importance, highlighting its impact on existing knowledge and potential applications. It addresses gaps, resolves problems, or contributes to advancements in a specific field. Emphasizing the study’s relevance, it demonstrates broader implications for society, academia, or industry, justifying the research effort and investment.
Significance of the Study Format
When writing the “Significance of the Study” section in a research paper , follow this format to ensure clarity and impact:
1. Introduction
- Contextual Background: Provide a brief background of the research topic.
- Research Problem: State the problem the study addresses.
2. Purpose of the Study
- Objective Statement: Clearly define the main objective of the study.
- Scope of the Study: Outline what the study covers.
3. Importance to the Field
- Contribution to Knowledge: Explain how the study will add to existing knowledge.
- Theoretical Significance: Discuss the study’s theoretical implications.
4. Practical Implications
- Real-world Application: Describe how the findings can be applied in practical setting .
- Beneficiaries: Identify who will benefit from the research (e.g., policymakers, practitioners, educators).
5. Advancement of Future Research
- Foundation for Future Studies: Indicate how the study can serve as a basis for further research.
- Research Gaps: Highlight any gaps the study aims to fill.
6. Societal Impact
- Broader Implications: Discuss the potential societal benefits or changes resulting from the study.
- Public Awareness: Explain how the study can raise awareness or understanding of the issue.
7. Conclusion
- Summary of Significance: Recap the main points that underline the importance of the study.
- Call to Action: Encourage specific actions or further studies based on the research findings.
Significance of the Study on Impact of Remote Work on Employee Productivity in the Tech Industry 1. Introduction The rapid shift to remote work due to the COVID-19 pandemic has fundamentally changed the dynamics of workplace productivity, especially within the tech industry. This study aims to examine how remote work influences employee productivity compared to traditional office settings. 2. Purpose of the Study The primary objective of this research is to evaluate the productivity levels of tech employees working remotely versus those working in office environments. The study analyzes various productivity metrics, such as task completion rates, quality of work, and employee satisfaction. 3. Importance to the Field This research contributes significantly to the existing body of knowledge by providing empirical data on the productivity impacts of remote work. It refines theoretical models of workplace productivity and offers new insights into remote work dynamics specific to the tech sector. Understanding these dynamics helps scholars and practitioners alike in shaping effective productivity strategies in the evolving work landscape. 4. Practical Implications The findings from this study have crucial practical implications for tech companies aiming to optimize their remote work policies. By understanding how remote work affects productivity, managers and HR departments can develop strategies to enhance employee performance and well-being in remote settings. These insights can also assist in designing training programs that equip employees with the skills needed for effective remote work. 5. Advancement of Future Research This study sets the stage for future research on long-term remote work trends and their impacts across various industries. It addresses existing gaps by providing a detailed analysis of how remote work influences productivity in the tech sector. Future researchers can build on this work to explore remote work dynamics in other fields and under different conditions. 6. Societal Impact The study highlights the broader societal implications of remote work, such as promoting work-life balance, reducing urban congestion, and lowering environmental pollution. By demonstrating the potential benefits of remote work, this research can influence public policy and corporate strategies towards more sustainable and flexible working conditions, ultimately contributing to societal well-being. 7. Conclusion Understanding the impact of remote work on productivity is essential for developing effective work policies and creating healthier work environments. This study provides valuable insights that can guide tech companies in optimizing their remote work strategies. Future research should explore the long-term effects of remote work across different sectors to provide a comprehensive understanding of its benefits and challenges.
Significance of the Study Examples
- Significance of the Study: Research Paper
- Significance of the Study: Qunatitive Research
- Significance of the Study: Qualitative Research
Research Paper

Qunatitive Research

Qualitative Research

More Significance of the Study Examples
- Educational Resources and Student Performance
- Business Innovation and Competitive Advantage
- Social Media Influencers and Brand Loyalty
- Mental Health Benefits of Physical Activ ity
- Sustainable Food Practices and Consumer Behavior
- Green Building and Energy Efficiency
- Technology in Healthcare
- Employee Engagement and Job Performance
- Business Strategies and Market Adaptation
- Mindfulness at Work
Purpose of Writing the Significance of a Study
When writing academic research or scholarly articles, one critical section is the significance of the study . This part addresses the importance and impact of the research, both theoretically and practically. Here are the main purposes of writing the significance of a study:
1. Establishing Relevance
The primary purpose is to explain why the study is relevant. It connects the research to existing literature, highlighting gaps or deficiencies that the current study aims to fill. This helps to justify the research problem and demonstrates the necessity of the study.
2. Highlighting Contributions
This section outlines the contributions the study will make to the field. It discusses how the findings can advance knowledge, theory, or practice. The significance emphasizes new insights, innovative approaches, or advancements that the study will provide.
3. Guiding Further Research
The significance of the study often includes suggestions for future research. By identifying limitations and unexplored areas, it encourages other researchers to pursue related questions. This helps to build a foundation for continuous inquiry and discovery.
4. Demonstrating Practical Applications
Beyond theoretical contributions, the significance of the study highlights practical applications. It shows how the research can solve real-world problems, improve practices, or influence policy-making. This connects academic research to practical outcomes that benefit society.
5. Engaging Stakeholders
Writing the significance of a study engages various stakeholders, including scholars, practitioners, policymakers, and funders. It communicates the value of the research to different audiences, making it easier to garner support, funding, or collaboration.
6. Enhancing Research Impact
A well-articulated significance section enhances the overall impact of the research. It underscores the importance and potential influence of the study, increasing its visibility and recognition in the academic community and beyond.
Benefits of Significance of the Study
Writing the significance of a study offers several benefits that enhance the research’s value and impact. Here are the key benefits:
1. Clarifies Research Value
The significance section clarifies the value of the research by explaining its importance and relevance. It helps readers understand why the study matters and what contributions it aims to make to the field.
2. Justifies the Research Problem
This section provides a rationale for the study by highlighting the research problem’s importance. It justifies the need for the study by identifying gaps in existing literature and explaining how the research will address these gaps.
3. Engages and Motivates Readers
A well-articulated significance section engages and motivates readers, including scholars, practitioners, and policymakers. It draws their interest by showcasing the study’s potential impact and benefits.
4. Secures Funding and Support
Explaining the significance of the study can help secure funding and support from stakeholders. Funding agencies and institutions are more likely to invest in research that demonstrates clear value and potential impact.
5. Guides Research Focus
The significance section helps guide the research focus by clearly defining the study’s contributions and goals. This clarity ensures that the research stays on track and aligns with its intended purpose.
6. Enhances Academic Credibility
Demonstrating the significance of a study enhances the researcher’s academic credibility. It shows a deep understanding of the field and the ability to identify and address important research questions.
7. Encourages Further Research
By identifying gaps and suggesting future research directions, the significance section encourages other researchers to build on the study’s findings. This fosters a continuous cycle of inquiry and discovery in the field.
8. Highlights Practical Applications
The significance section highlights practical applications of the research, showing how it can solve real-world problems. This makes the study more appealing to practitioners and policymakers who are interested in practical solutions.
9. Increases Research Impact
A clear and compelling significance section increases the overall impact of the research. It enhances the study’s visibility and recognition, leading to broader dissemination and application of the findings.
10. Supports Academic and Professional Goals
For researchers, writing a strong significance section supports academic and professional goals. It can contribute to career advancement, publication opportunities, and recognition within the academic community.
How to Write the Significance of the Study

Writing the significance of a study involves explaining the importance and impact of your research. This section should clearly articulate why your study matters, how it contributes to the field, and what practical applications it may have. Here’s a step-by-step guide to help you write an effective significance of the study:
Start with the Context
Begin by providing a brief overview of the research context. This sets the stage for understanding the importance of your study. Example : “In today’s digital age, digital literacy has become a critical skill for students. As technology continues to integrate into education, understanding its impact on academic performance is essential.”
Identify the Research Gap
Explain the gap in existing literature or the problem your study aims to address. Highlighting this gap justifies the need for your research. Example: “Despite the growing importance of digital literacy, there is limited empirical evidence on its direct impact on high school students’ academic performance. This study seeks to fill this gap by investigating this relationship.”
Explain the Theoretical Contributions
Discuss how your study will contribute to existing theories or knowledge in the field. This shows the academic value of your research. Example : “The findings of this study will contribute to educational theory by providing new insights into how digital literacy skills influence student learning outcomes. It will expand the current understanding of the role of technology in education.”
Highlight Practical Implications
Describe the practical applications of your research. Explain how the findings can be used in real-world settings. Example : “Practically, the results of this study can inform educators and policymakers about the importance of incorporating digital literacy programs into the curriculum. It will help design more effective teaching strategies that enhance students’ digital competencies.”
Mention the Beneficiaries
Identify who will benefit from your study. This could include scholars, practitioners, policymakers, or specific groups affected by the research problem. Example: “This research will benefit educators, school administrators, and policymakers by providing evidence-based recommendations for integrating digital literacy into educational practices. Additionally, students will benefit from improved learning outcomes and better preparedness for the digital world.”
Suggest Future Research
Point out areas for future research that stem from your study. This shows the ongoing relevance and potential for further inquiry. Example : “Future research could explore the long-term effects of digital literacy on career readiness and job performance. Additionally, studies could examine the impact of specific digital literacy interventions on diverse student populations.”
Use Clear and Concise Language
Ensure your writing is clear and concise. Avoid jargon and overly complex sentences to make your significance easily understandable.
What is the significance of a study?
The significance explains the importance, contributions, and impact of the research, highlighting why the study is necessary and how it benefits the field and society.
Why is the significance of a study important?
It justifies the research, engages readers, secures funding, guides the research focus, and highlights practical and theoretical contributions, enhancing the study’s impact and visibility.
How do you identify the significance of a study?
Identify gaps in existing literature, potential contributions to theory and practice, and practical applications that address real-world problems, demonstrating the study’s relevance and importance.
What should be included in the significance of a study?
Include the research context, identified gaps, theoretical contributions, practical applications, beneficiaries, and suggestions for future research to comprehensively explain the study’s importance.
How long should the significance of a study be?
Typically, the significance section should be concise, around 1-2 paragraphs, providing enough detail to clearly convey the study’s importance and contributions.
Can the significance of a study influence funding decisions?
Yes, a well-articulated significance section can attract funding by demonstrating the study’s potential impact and relevance to funding agencies and stakeholders.
How does the significance of a study benefit researchers?
It clarifies the research focus, enhances credibility, guides the research process, and supports academic and professional goals by highlighting the study’s contributions and importance.
Should the significance of a study mention future research?
Yes, mentioning future research directions shows the ongoing relevance of the study and encourages further inquiry, contributing to continuous advancement in the field.
How does the significance of a study relate to the research problem?
The significance justifies the research problem by explaining its importance, highlighting gaps in existing knowledge, and showing how the study addresses these issues.
Can practical applications be part of the significance of a study?
Yes, practical applications are crucial, showing how the research can solve real-world problems, influence practices, and benefit specific groups or society overall.
Text prompt
- Instructive
- Professional
10 Examples of Public speaking
20 Examples of Gas lighting

Community Blog
Keep up-to-date on postgraduate related issues with our quick reads written by students, postdocs, professors and industry leaders.
What is the Significance of the Study?
- By DiscoverPhDs
- August 25, 2020

- what the significance of the study means,
- why it’s important to include in your research work,
- where you would include it in your paper, thesis or dissertation,
- how you write one
- and finally an example of a well written section about the significance of the study.
What does Significance of the Study mean?
The significance of the study is a written statement that explains why your research was needed. It’s a justification of the importance of your work and impact it has on your research field, it’s contribution to new knowledge and how others will benefit from it.
Why is the Significance of the Study important?
The significance of the study, also known as the rationale of the study, is important to convey to the reader why the research work was important. This may be an academic reviewer assessing your manuscript under peer-review, an examiner reading your PhD thesis, a funder reading your grant application or another research group reading your published journal paper. Your academic writing should make clear to the reader what the significance of the research that you performed was, the contribution you made and the benefits of it.
How do you write the Significance of the Study?
When writing this section, first think about where the gaps in knowledge are in your research field. What are the areas that are poorly understood with little or no previously published literature? Or what topics have others previously published on that still require further work. This is often referred to as the problem statement.
The introduction section within the significance of the study should include you writing the problem statement and explaining to the reader where the gap in literature is.
Then think about the significance of your research and thesis study from two perspectives: (1) what is the general contribution of your research on your field and (2) what specific contribution have you made to the knowledge and who does this benefit the most.
For example, the gap in knowledge may be that the benefits of dumbbell exercises for patients recovering from a broken arm are not fully understood. You may have performed a study investigating the impact of dumbbell training in patients with fractures versus those that did not perform dumbbell exercises and shown there to be a benefit in their use. The broad significance of the study would be the improvement in the understanding of effective physiotherapy methods. Your specific contribution has been to show a significant improvement in the rate of recovery in patients with broken arms when performing certain dumbbell exercise routines.
This statement should be no more than 500 words in length when written for a thesis. Within a research paper, the statement should be shorter and around 200 words at most.
Significance of the Study: An example
Building on the above hypothetical academic study, the following is an example of a full statement of the significance of the study for you to consider when writing your own. Keep in mind though that there’s no single way of writing the perfect significance statement and it may well depend on the subject area and the study content.
Here’s another example to help demonstrate how a significance of the study can also be applied to non-technical fields:
The significance of this research lies in its potential to inform clinical practices and patient counseling. By understanding the psychological outcomes associated with non-surgical facial aesthetics, practitioners can better guide their patients in making informed decisions about their treatment plans. Additionally, this study contributes to the body of academic knowledge by providing empirical evidence on the effects of these cosmetic procedures, which have been largely anecdotal up to this point.
The statement of the significance of the study is used by students and researchers in academic writing to convey the importance of the research performed; this section is written at the end of the introduction and should describe the specific contribution made and who it benefits.

Learning how to effectively collaborate with others is an important skill for anyone in academia to develop.

A thesis and dissertation appendix contains additional information which supports your main arguments. Find out what they should include and how to format them.

Find out the differences between a Literature Review and an Annotated Bibliography, whey they should be used and how to write them.
Join thousands of other students and stay up to date with the latest PhD programmes, funding opportunities and advice.

Browse PhDs Now

In the UK, a dissertation, usually around 20,000 words is written by undergraduate and Master’s students, whilst a thesis, around 80,000 words, is written as part of a PhD.

The Thurstone Scale is used to quantify the attitudes of people being surveyed, using a format of ‘agree-disagree’ statements.

Priya’s a 1st year PhD student University College Dublin. Her project involves investigating a novel seaweed-ensiling process as an alternative to drying to preserve seaweeds nutritional and monetary value.

Gabrielle’s a 2nd year Immunology PhD student at the University of Michigan. Her research focus on the complications of obesity and type 2 diabetes in the clearance of respiratory bacterial infections.
Join Thousands of Students
How To Write a Significance Statement for Your Research
A significance statement is an essential part of a research paper. It explains the importance and relevance of the study to the academic community and the world at large. To write a compelling significance statement, identify the research problem, explain why it is significant, provide evidence of its importance, and highlight its potential impact on future research, policy, or practice. A well-crafted significance statement should effectively communicate the value of the research to readers and help them understand why it matters.
Updated on May 4, 2023

A significance statement is a clearly stated, non-technical paragraph that explains why your research matters. It’s central in making the public aware of and gaining support for your research.
Write it in jargon-free language that a reader from any field can understand. Well-crafted, easily readable significance statements can improve your chances for citation and impact and make it easier for readers outside your field to find and understand your work.
Read on for more details on what a significance statement is, how it can enhance the impact of your research, and, of course, how to write one.
What is a significance statement in research?
A significance statement answers the question: How will your research advance scientific knowledge and impact society at large (as well as specific populations)?
You might also see it called a “Significance of the study” statement. Some professional organizations in the STEM sciences and social sciences now recommended that journals in their disciplines make such statements a standard feature of each published article. Funding agencies also consider “significance” a key criterion for their awards.
Read some examples of significance statements from the Proceedings of the National Academy of Sciences (PNAS) here .
Depending upon the specific journal or funding agency’s requirements, your statement may be around 100 words and answer these questions:
1. What’s the purpose of this research?
2. What are its key findings?
3. Why do they matter?
4. Who benefits from the research results?
Readers will want to know: “What is interesting or important about this research?” Keep asking yourself that question.
Where to place the significance statement in your manuscript
Most journals ask you to place the significance statement before or after the abstract, so check with each journal’s guide.
This article is focused on the formal significance statement, even though you’ll naturally highlight your project’s significance elsewhere in your manuscript. (In the introduction, you’ll set out your research aims, and in the conclusion, you’ll explain the potential applications of your research and recommend areas for future research. You’re building an overall case for the value of your work.)
Developing the significance statement
The main steps in planning and developing your statement are to assess the gaps to which your study contributes, and then define your work’s implications and impact.
Identify what gaps your study fills and what it contributes
Your literature review was a big part of how you planned your study. To develop your research aims and objectives, you identified gaps or unanswered questions in the preceding research and designed your study to address them.
Go back to that lit review and look at those gaps again. Review your research proposal to refresh your memory. Ask:
- How have my research findings advanced knowledge or provided notable new insights?
- How has my research helped to prove (or disprove) a hypothesis or answer a research question?
- Why are those results important?
Consider your study’s potential impact at two levels:
- What contribution does my research make to my field?
- How does it specifically contribute to knowledge; that is, who will benefit the most from it?
Define the implications and potential impact
As you make notes, keep the reasons in mind for why you are writing this statement. Whom will it impact, and why?
The first audience for your significance statement will be journal reviewers when you submit your article for publishing. Many journals require one for manuscript submissions. Study the author’s guide of your desired journal to see its criteria ( here’s an example ). Peer reviewers who can clearly understand the value of your research will be more likely to recommend publication.
Second, when you apply for funding, your significance statement will help justify why your research deserves a grant from a funding agency . The U.S. National Institutes of Health (NIH), for example, wants to see that a project will “exert a sustained, powerful influence on the research field(s) involved.” Clear, simple language is always valuable because not all reviewers will be specialists in your field.
Third, this concise statement about your study’s importance can affect how potential readers engage with your work. Science journalists and interested readers can promote and spread your work, enhancing your reputation and influence. Help them understand your work.
You’re now ready to express the importance of your research clearly and concisely. Time to start writing.
How to write a significance statement: Key elements
When drafting your statement, focus on both the content and writing style.
- In terms of content, emphasize the importance, timeliness, and relevance of your research results.
- Write the statement in plain, clear language rather than scientific or technical jargon. Your audience will include not just your fellow scientists but also non-specialists like journalists, funding reviewers, and members of the public.
Follow the process we outline below to build a solid, well-crafted, and informative statement.
Get started
Some suggested opening lines to help you get started might be:
- The implications of this study are…
- Building upon previous contributions, our study moves the field forward because…
- Our study furthers previous understanding about…
Alternatively, you may start with a statement about the phenomenon you’re studying, leading to the problem statement.
Include these components
Next, draft some sentences that include the following elements. A good example, which we’ll use here, is a significance statement by Rogers et al. (2022) published in the Journal of Climate .
1. Briefly situate your research study in its larger context . Start by introducing the topic, leading to a problem statement. Here’s an example:
‘Heatwaves pose a major threat to human health, ecosystems, and human systems.”
2. State the research problem.
“Simultaneous heatwaves affecting multiple regions can exacerbate such threats. For example, multiple food-producing regions simultaneously undergoing heat-related crop damage could drive global food shortages.”
3. Tell what your study does to address it.
“We assess recent changes in the occurrence of simultaneous large heatwaves.”
4. Provide brief but powerful evidence to support the claims your statement is making , Use quantifiable terms rather than vague ones (e.g., instead of “This phenomenon is happening now more than ever,” see below how Rogers et al. (2022) explained it). This evidence intensifies and illustrates the problem more vividly:
“Such simultaneous heatwaves are 7 times more likely now than 40 years ago. They are also hotter and affect a larger area. Their increasing occurrence is mainly driven by warming baseline temperatures due to global heating, but changes in weather patterns contribute to disproportionate increases over parts of Europe, the eastern United States, and Asia.
5. Relate your study’s impact to the broader context , starting with its general significance to society—then, when possible, move to the particular as you name specific applications of your research findings. (Our example lacks this second level of application.)
“Better understanding the drivers of weather pattern changes is therefore important for understanding future concurrent heatwave characteristics and their impacts.”
Refine your English
Don’t understate or overstate your findings – just make clear what your study contributes. When you have all the elements in place, review your draft to simplify and polish your language. Even better, get an expert AJE edit . Be sure to use “plain” language rather than academic jargon.
- Avoid acronyms, scientific jargon, and technical terms
- Use active verbs in your sentence structure rather than passive voice (e.g., instead of “It was found that...”, use “We found...”)
- Make sentence structures short, easy to understand – readable
- Try to address only one idea in each sentence and keep sentences within 25 words (15 words is even better)
- Eliminate nonessential words and phrases (“fluff” and wordiness)
Enhance your significance statement’s impact
Always take time to review your draft multiple times. Make sure that you:
- Keep your language focused
- Provide evidence to support your claims
- Relate the significance to the broader research context in your field
After revising your significance statement, request feedback from a reading mentor about how to make it even clearer. If you’re not a native English speaker, seek help from a native-English-speaking colleague or use an editing service like AJE to make sure your work is at a native level.
Understanding the significance of your study
Your readers may have much less interest than you do in the specific details of your research methods and measures. Many readers will scan your article to learn how your findings might apply to them and their own research.
Different types of significance
Your findings may have different types of significance, relevant to different populations or fields of study for different reasons. You can emphasize your work’s statistical, clinical, or practical significance. Editors or reviewers in the social sciences might also evaluate your work’s social or political significance.
Statistical significance means that the results are unlikely to have occurred randomly. Instead, it implies a true cause-and-effect relationship.
Clinical significance means that your findings are applicable for treating patients and improving quality of life.
Practical significance is when your research outcomes are meaningful to society at large, in the “real world.” Practical significance is usually measured by the study’s effect size . Similarly, evaluators may attribute social or political significance to research that addresses “real and immediate” social problems.

The AJE Team
See our "Privacy Policy"
Home » Feature » Thesis » Significance of the Study Samples | Writing Tips
Significance of the Study Samples | Writing Tips
When you write a thesis , there is a section there that is allocated for the significance of the study. This article will provide different significance of the study examples and will discuss tips on how to write this part.
Tips in Writing the Significance of the Study
Here are the tips that may be helpful when writing the significance of the study. These tips will tell you the basic components expected to be seen in the significance of the study content.
1. Refer to the Problem Statement
In writing the significance of the study, always refer to the statement of the problem. This way, you can clearly define the contribution of your study. To simplify, your research should answer this question, “What are the benefits or advantages of the study based on the statement of the problem?”
Start by explaining the problem that your study aimed to solve. For example, if you conducted a research study on obesity rates among elementary school students, you would start by explaining that obesity is a major health concern in the Philippines and discuss why it is important to find ways to address this issue.
2. Write it from General to Particular
Determine the specific contribution of your thesis study to society as well as to the individual. Write it deductively, starting from general to specific. Start your significance of the study broadly then narrow it out to a specific group or person. This is done by looking into the general contribution of your study, such as its importance to society as a whole, then moving towards its contribution to individuals like yourself as a researcher.
Discuss how your study fills a gap in the literature. If you conducted an experiment on the effects of a certain type of food on children, for example, you might start by explaining that no research has been done on this topic before. This section would also include a discussion about why your study is important.
Your problem statement might help you determine the unique contribution of your research. This can be accomplished by ensuring that the aim of the problem and the study’s objectives are identical. For instance, if your research question is “Is there a significant relationship between the use of Facebook Messenger and the performance of students in English spelling? “, you could write as one of the contributions of your study: “The study will identify common errors in spelling and grammar by Messenger users and recommend its appropriate use in a way that can improve performance in spelling.”
You may also read: How to Make a Conceptual Framework
Significance of the Study Samples
Here are some examples to help you draft your own introduction:
Title: Number of Clinical Internship Hours: A Determinant of Student’s Effectiveness and Skill Acquisition in the Hospital Area for Velez College Students
Significance of the study.
The results of the study will be of great benefit to the following:
College of Nursing Dean . Data given will provide the dean with information on how the number of duty hours in a week affects the student’s academic and RLE performance. The results will enable the dean to improve the scheduling of RLE and different academic subjects. Data gathered will help the dean initiate collaboration among faculty and chairpersons to help plan the advancement of nursing education in relation to the new curriculum.
Clinical Instructors . The results of the study will help the clinical instructors evaluate the quality of care rendered by the nursing students, academic performance, attitude and skills acquired in relation to the number of hours given in a week. Results would also develop the clinical instructor’s teaching-learning and evaluating strategies in enhancing knowledge, skills and attitude to the students in the time frame given.
Students . This study will provide information regarding which time arrangement is effective: 8-hr of clinical internship from the 5-hr clinical internship with additional academic classes. This study will evaluate the academic performance, the student nurse’s attitude and approach, the skills learned in the clinical area, and the quality of care rendered in the given time frame. Data gathered will also help the students improve both academic and clinical performance.
Velez College . This study will improve the school in the development of nursing education. This study will foster new ways of enhancing knowledge, skills, and attitude, thus preparing globally-competitive nurses in the future. This study will also help in the advancement of school management, clinical leadership, and the teaching-evaluation approach.
Title: The Effectiveness of Isuzu’s Blue Power Technology in Fuel Efficiency of Diesel Engines
The generalization of this study would be a great contribution to the vast knowledge in relation to the brand awareness of Isuzu’s Blue Power Euro 4 Technology. Furthermore, the results of this investigation could be highly significant and beneficial for the following:
Current Customers
They refer to consumers that have already bought products from Isuzu. They are considered to be the main beneficiaries of the business. The findings of this study would provide them with adequate information about the product, most especially for those clients that have already bought units with the Blue Power Euro 4 Technology but have no idea of its benefits and advantages.
Potential Customer
They are the consumers that have not yet purchased this brand. This study aims to give them insights and overviews of the product and would help them choose the right variant to purchase.
They are the main beneficiaries of this study, which may help them to improve their marketing strategies. It would provide substantial data to the business that they could make use of in boosting their sales. Moreover, developing brand awareness will cater to more demands and loyalty in the future.
For they also play a vital role in the business and as consumers. This research would give them the idea that such private vehicles exist, which helps them to conserve energy rather than exploit it. Hence, giving back to the community and making it a better place to live.
Proponents of the Study
This refers to the students conducting the study. They will find self-fulfillment and gain knowledge and skills in this study. This study will help and inspire more researchers to be more innovative and creative in their future endeavors.
Future Researchers
This study will serve as a reference for researchers on the subject of research in the field of marketing. This will serve as a guide to further developing the research with the connection to the variables used.
The significance of a study is a key component of a strong scientific paper. By following these tips, you can create a clear and concise explanation of the importance of your work. I hope that these tips and samples will help you create a perfect Significance of the Study for your thesis. Apply these tips to prevent your mind from wandering aimlessly as you draft the significance of the study. It will allow you to focus on the next section of your thesis, helping you finish it on time. Good luck!
good and interesting
thanks for information
Very useful. Thanks
Thank you for a very informative article
- Civil Service Exam Reviewer
- L TO Portal Registration Guide
- How to Renew a Driver's License
- National ID Tracker
- Civil Service Exam Application
- Privacy Policy
- Terms of Service
All content provided on this website is for informational purposes only. The owner of this website makes no representations as to the accuracy or completeness of any information on this site or found by following any link on this site. The owner will not be liable for any errors or omissions in this information or for the availability of this information. The owner will not be liable for any losses, injuries, or damages from the display or use of this information.
You may not use the content of this blog for commercial purposes without prior formal written consent from us. These terms and conditions of use are subject to change at any time and without notice.
TOPNOTCHER PH is a participant in the Shopee Affiliate Program, an affiliate advertising program designed to provide a means for us to earn advertising fees by linking to SHOPEE.PH. As a Shopee Affiliate, we earn from your qualifying purchases through our links without extra cost to you.
© 2024 TOPNOTCHER.PH
- Privacy Policy

Home » Background of The Study – Examples and Writing Guide
Background of The Study – Examples and Writing Guide
Table of Contents

Background of The Study
Definition:
Background of the study refers to the context, circumstances, and history that led to the research problem or topic being studied. It provides the reader with a comprehensive understanding of the subject matter and the significance of the study.
The background of the study usually includes a discussion of the relevant literature, the gap in knowledge or understanding, and the research questions or hypotheses to be addressed. It also highlights the importance of the research topic and its potential contributions to the field. A well-written background of the study sets the stage for the research and helps the reader to appreciate the need for the study and its potential significance.
How to Write Background of The Study
Here are some steps to help you write the background of the study:
Identify the Research Problem
Start by identifying the research problem you are trying to address. This problem should be significant and relevant to your field of study.
Provide Context
Once you have identified the research problem, provide some context. This could include the historical, social, or political context of the problem.
Review Literature
Conduct a thorough review of the existing literature on the topic. This will help you understand what has been studied and what gaps exist in the current research.
Identify Research Gap
Based on your literature review, identify the gap in knowledge or understanding that your research aims to address. This gap will be the focus of your research question or hypothesis.
State Objectives
Clearly state the objectives of your research . These should be specific, measurable, achievable, relevant, and time-bound (SMART).
Discuss Significance
Explain the significance of your research. This could include its potential impact on theory , practice, policy, or society.
Finally, summarize the key points of the background of the study. This will help the reader understand the research problem, its context, and its significance.
How to Write Background of The Study in Proposal
The background of the study is an essential part of any proposal as it sets the stage for the research project and provides the context and justification for why the research is needed. Here are the steps to write a compelling background of the study in your proposal:
- Identify the problem: Clearly state the research problem or gap in the current knowledge that you intend to address through your research.
- Provide context: Provide a brief overview of the research area and highlight its significance in the field.
- Review literature: Summarize the relevant literature related to the research problem and provide a critical evaluation of the current state of knowledge.
- Identify gaps : Identify the gaps or limitations in the existing literature and explain how your research will contribute to filling these gaps.
- Justify the study : Explain why your research is important and what practical or theoretical contributions it can make to the field.
- Highlight objectives: Clearly state the objectives of the study and how they relate to the research problem.
- Discuss methodology: Provide an overview of the methodology you will use to collect and analyze data, and explain why it is appropriate for the research problem.
- Conclude : Summarize the key points of the background of the study and explain how they support your research proposal.
How to Write Background of The Study In Thesis
The background of the study is a critical component of a thesis as it provides context for the research problem, rationale for conducting the study, and the significance of the research. Here are some steps to help you write a strong background of the study:
- Identify the research problem : Start by identifying the research problem that your thesis is addressing. What is the issue that you are trying to solve or explore? Be specific and concise in your problem statement.
- Review the literature: Conduct a thorough review of the relevant literature on the topic. This should include scholarly articles, books, and other sources that are directly related to your research question.
- I dentify gaps in the literature: After reviewing the literature, identify any gaps in the existing research. What questions remain unanswered? What areas have not been explored? This will help you to establish the need for your research.
- Establish the significance of the research: Clearly state the significance of your research. Why is it important to address this research problem? What are the potential implications of your research? How will it contribute to the field?
- Provide an overview of the research design: Provide an overview of the research design and methodology that you will be using in your study. This should include a brief explanation of the research approach, data collection methods, and data analysis techniques.
- State the research objectives and research questions: Clearly state the research objectives and research questions that your study aims to answer. These should be specific, measurable, achievable, relevant, and time-bound.
- Summarize the chapter: Summarize the chapter by highlighting the key points and linking them back to the research problem, significance of the study, and research questions.
How to Write Background of The Study in Research Paper
Here are the steps to write the background of the study in a research paper:
- Identify the research problem: Start by identifying the research problem that your study aims to address. This can be a particular issue, a gap in the literature, or a need for further investigation.
- Conduct a literature review: Conduct a thorough literature review to gather information on the topic, identify existing studies, and understand the current state of research. This will help you identify the gap in the literature that your study aims to fill.
- Explain the significance of the study: Explain why your study is important and why it is necessary. This can include the potential impact on the field, the importance to society, or the need to address a particular issue.
- Provide context: Provide context for the research problem by discussing the broader social, economic, or political context that the study is situated in. This can help the reader understand the relevance of the study and its potential implications.
- State the research questions and objectives: State the research questions and objectives that your study aims to address. This will help the reader understand the scope of the study and its purpose.
- Summarize the methodology : Briefly summarize the methodology you used to conduct the study, including the data collection and analysis methods. This can help the reader understand how the study was conducted and its reliability.
Examples of Background of The Study
Here are some examples of the background of the study:
Problem : The prevalence of obesity among children in the United States has reached alarming levels, with nearly one in five children classified as obese.
Significance : Obesity in childhood is associated with numerous negative health outcomes, including increased risk of type 2 diabetes, cardiovascular disease, and certain cancers.
Gap in knowledge : Despite efforts to address the obesity epidemic, rates continue to rise. There is a need for effective interventions that target the unique needs of children and their families.
Problem : The use of antibiotics in agriculture has contributed to the development of antibiotic-resistant bacteria, which poses a significant threat to human health.
Significance : Antibiotic-resistant infections are responsible for thousands of deaths each year and are a major public health concern.
Gap in knowledge: While there is a growing body of research on the use of antibiotics in agriculture, there is still much to be learned about the mechanisms of resistance and the most effective strategies for reducing antibiotic use.
Edxample 3:
Problem : Many low-income communities lack access to healthy food options, leading to high rates of food insecurity and diet-related diseases.
Significance : Poor nutrition is a major contributor to chronic diseases such as obesity, type 2 diabetes, and cardiovascular disease.
Gap in knowledge : While there have been efforts to address food insecurity, there is a need for more research on the barriers to accessing healthy food in low-income communities and effective strategies for increasing access.
Examples of Background of The Study In Research
Here are some real-life examples of how the background of the study can be written in different fields of study:
Example 1 : “There has been a significant increase in the incidence of diabetes in recent years. This has led to an increased demand for effective diabetes management strategies. The purpose of this study is to evaluate the effectiveness of a new diabetes management program in improving patient outcomes.”
Example 2 : “The use of social media has become increasingly prevalent in modern society. Despite its popularity, little is known about the effects of social media use on mental health. This study aims to investigate the relationship between social media use and mental health in young adults.”
Example 3: “Despite significant advancements in cancer treatment, the survival rate for patients with pancreatic cancer remains low. The purpose of this study is to identify potential biomarkers that can be used to improve early detection and treatment of pancreatic cancer.”
Examples of Background of The Study in Proposal
Here are some real-time examples of the background of the study in a proposal:
Example 1 : The prevalence of mental health issues among university students has been increasing over the past decade. This study aims to investigate the causes and impacts of mental health issues on academic performance and wellbeing.
Example 2 : Climate change is a global issue that has significant implications for agriculture in developing countries. This study aims to examine the adaptive capacity of smallholder farmers to climate change and identify effective strategies to enhance their resilience.
Example 3 : The use of social media in political campaigns has become increasingly common in recent years. This study aims to analyze the effectiveness of social media campaigns in mobilizing young voters and influencing their voting behavior.
Example 4 : Employee turnover is a major challenge for organizations, especially in the service sector. This study aims to identify the key factors that influence employee turnover in the hospitality industry and explore effective strategies for reducing turnover rates.
Examples of Background of The Study in Thesis
Here are some real-time examples of the background of the study in the thesis:
Example 1 : “Women’s participation in the workforce has increased significantly over the past few decades. However, women continue to be underrepresented in leadership positions, particularly in male-dominated industries such as technology. This study aims to examine the factors that contribute to the underrepresentation of women in leadership roles in the technology industry, with a focus on organizational culture and gender bias.”
Example 2 : “Mental health is a critical component of overall health and well-being. Despite increased awareness of the importance of mental health, there are still significant gaps in access to mental health services, particularly in low-income and rural communities. This study aims to evaluate the effectiveness of a community-based mental health intervention in improving mental health outcomes in underserved populations.”
Example 3: “The use of technology in education has become increasingly widespread, with many schools adopting online learning platforms and digital resources. However, there is limited research on the impact of technology on student learning outcomes and engagement. This study aims to explore the relationship between technology use and academic achievement among middle school students, as well as the factors that mediate this relationship.”
Examples of Background of The Study in Research Paper
Here are some examples of how the background of the study can be written in various fields:
Example 1: The prevalence of obesity has been on the rise globally, with the World Health Organization reporting that approximately 650 million adults were obese in 2016. Obesity is a major risk factor for several chronic diseases such as diabetes, cardiovascular diseases, and cancer. In recent years, several interventions have been proposed to address this issue, including lifestyle changes, pharmacotherapy, and bariatric surgery. However, there is a lack of consensus on the most effective intervention for obesity management. This study aims to investigate the efficacy of different interventions for obesity management and identify the most effective one.
Example 2: Antibiotic resistance has become a major public health threat worldwide. Infections caused by antibiotic-resistant bacteria are associated with longer hospital stays, higher healthcare costs, and increased mortality. The inappropriate use of antibiotics is one of the main factors contributing to the development of antibiotic resistance. Despite numerous efforts to promote the rational use of antibiotics, studies have shown that many healthcare providers continue to prescribe antibiotics inappropriately. This study aims to explore the factors influencing healthcare providers’ prescribing behavior and identify strategies to improve antibiotic prescribing practices.
Example 3: Social media has become an integral part of modern communication, with millions of people worldwide using platforms such as Facebook, Twitter, and Instagram. Social media has several advantages, including facilitating communication, connecting people, and disseminating information. However, social media use has also been associated with several negative outcomes, including cyberbullying, addiction, and mental health problems. This study aims to investigate the impact of social media use on mental health and identify the factors that mediate this relationship.
Purpose of Background of The Study
The primary purpose of the background of the study is to help the reader understand the rationale for the research by presenting the historical, theoretical, and empirical background of the problem.
More specifically, the background of the study aims to:
- Provide a clear understanding of the research problem and its context.
- Identify the gap in knowledge that the study intends to fill.
- Establish the significance of the research problem and its potential contribution to the field.
- Highlight the key concepts, theories, and research findings related to the problem.
- Provide a rationale for the research questions or hypotheses and the research design.
- Identify the limitations and scope of the study.
When to Write Background of The Study
The background of the study should be written early on in the research process, ideally before the research design is finalized and data collection begins. This allows the researcher to clearly articulate the rationale for the study and establish a strong foundation for the research.
The background of the study typically comes after the introduction but before the literature review section. It should provide an overview of the research problem and its context, and also introduce the key concepts, theories, and research findings related to the problem.
Writing the background of the study early on in the research process also helps to identify potential gaps in knowledge and areas for further investigation, which can guide the development of the research questions or hypotheses and the research design. By establishing the significance of the research problem and its potential contribution to the field, the background of the study can also help to justify the research and secure funding or support from stakeholders.
Advantage of Background of The Study
The background of the study has several advantages, including:
- Provides context: The background of the study provides context for the research problem by highlighting the historical, theoretical, and empirical background of the problem. This allows the reader to understand the research problem in its broader context and appreciate its significance.
- Identifies gaps in knowledge: By reviewing the existing literature related to the research problem, the background of the study can identify gaps in knowledge that the study intends to fill. This helps to establish the novelty and originality of the research and its potential contribution to the field.
- Justifies the research : The background of the study helps to justify the research by demonstrating its significance and potential impact. This can be useful in securing funding or support for the research.
- Guides the research design: The background of the study can guide the development of the research questions or hypotheses and the research design by identifying key concepts, theories, and research findings related to the problem. This ensures that the research is grounded in existing knowledge and is designed to address the research problem effectively.
- Establishes credibility: By demonstrating the researcher’s knowledge of the field and the research problem, the background of the study can establish the researcher’s credibility and expertise, which can enhance the trustworthiness and validity of the research.
Disadvantages of Background of The Study
Some Disadvantages of Background of The Study are as follows:
- Time-consuming : Writing a comprehensive background of the study can be time-consuming, especially if the research problem is complex and multifaceted. This can delay the research process and impact the timeline for completing the study.
- Repetitive: The background of the study can sometimes be repetitive, as it often involves summarizing existing research and theories related to the research problem. This can be tedious for the reader and may make the section less engaging.
- Limitations of existing research: The background of the study can reveal the limitations of existing research related to the problem. This can create challenges for the researcher in developing research questions or hypotheses that address the gaps in knowledge identified in the background of the study.
- Bias : The researcher’s biases and perspectives can influence the content and tone of the background of the study. This can impact the reader’s perception of the research problem and may influence the validity of the research.
- Accessibility: Accessing and reviewing the literature related to the research problem can be challenging, especially if the researcher does not have access to a comprehensive database or if the literature is not available in the researcher’s language. This can limit the depth and scope of the background of the study.
About the author
Muhammad Hassan
Researcher, Academic Writer, Web developer
You may also like

Research Techniques – Methods, Types and Examples

Thesis – Structure, Example and Writing Guide

Chapter Summary & Overview – Writing Guide...

Research Problem – Examples, Types and Guide

Research Report – Example, Writing Guide and...

Informed Consent in Research – Types, Templates...
- Link to facebook
- Link to linkedin
- Link to twitter
- Link to youtube
- Writing Tips
How to Discuss the Significance of Your Research

6-minute read
- 10th April 2023
Introduction
Research papers can be a real headache for college students . As a student, your research needs to be credible enough to support your thesis statement. You must also ensure you’ve discussed the literature review, findings, and results.
However, it’s also important to discuss the significance of your research . Your potential audience will care deeply about this. It will also help you conduct your research. By knowing the impact of your research, you’ll understand what important questions to answer.
If you’d like to know more about the impact of your research, read on! We’ll talk about why it’s important and how to discuss it in your paper.
What Is the Significance of Research?
This is the potential impact of your research on the field of study. It includes contributions from new knowledge from the research and those who would benefit from it. You should present this before conducting research, so you need to be aware of current issues associated with the thesis before discussing the significance of the research.
Why Does the Significance of Research Matter?
Potential readers need to know why your research is worth pursuing. Discussing the significance of research answers the following questions:
● Why should people read your research paper ?
● How will your research contribute to the current knowledge related to your topic?
● What potential impact will it have on the community and professionals in the field?
Not including the significance of research in your paper would be like a knight trying to fight a dragon without weapons.
Where Do I Discuss the Significance of Research in My Paper?
As previously mentioned, the significance of research comes before you conduct it. Therefore, you should discuss the significance of your research in the Introduction section. Your reader should know the problem statement and hypothesis beforehand.

Steps to Discussing the Significance of Your Research
Discussing the significance of research might seem like a loaded question, so we’ve outlined some steps to help you tackle it.
Step 1: The Research Problem
The problem statement can reveal clues about the outcome of your research. Your research should provide answers to the problem, which is beneficial to all those concerned. For example, imagine the problem statement is, “To what extent do elementary and high school teachers believe cyberbullying affects student performance?”
Learning teachers’ opinions on the effects of cyberbullying on student performance could result in the following:
● Increased public awareness of cyberbullying in elementary and high schools
● Teachers’ perceptions of cyberbullying negatively affecting student performance
Find this useful?
Subscribe to our newsletter and get writing tips from our editors straight to your inbox.
● Whether cyberbullying is more prevalent in elementary or high schools
The research problem will steer your research in the right direction, so it’s best to start with the problem statement.
Step 2: Existing Literature in the Field
Think about current information on your topic, and then find out what information is missing. Are there any areas that haven’t been explored? Your research should add new information to the literature, so be sure to state this in your discussion. You’ll need to know the current literature on your topic anyway, as this is part of your literature review section .
Step 3: Your Research’s Impact on Society
Inform your readers about the impact on society your research could have on it. For example, in the study about teachers’ opinions on cyberbullying, you could mention that your research will educate the community about teachers’ perceptions of cyberbullying as it affects student performance. As a result, the community will know how many teachers believe cyberbullying affects student performance.
You can also mention specific individuals and institutions that would benefit from your study. In the example of cyberbullying, you might indicate that school principals and superintendents would benefit from your research.
Step 4: Future Studies in the Field
Next, discuss how the significance of your research will benefit future studies, which is especially helpful for future researchers in your field. In the example of cyberbullying affecting student performance, your research could provide further opportunities to assess teacher perceptions of cyberbullying and its effects on students from larger populations. This prepares future researchers for data collection and analysis.
Discussing the significance of your research may sound daunting when you haven’t conducted it yet. However, an audience might not read your paper if they don’t know the significance of the research. By focusing on the problem statement and the research benefits to society and future studies, you can convince your audience of the value of your research.
Remember that everything you write doesn’t have to be set in stone. You can go back and tweak the significance of your research after conducting it. At first, you might only include general contributions of your study, but as you research, your contributions will become more specific.
You should have a solid understanding of your topic in general, its associated problems, and the literature review before tackling the significance of your research. However, you’re not trying to prove your thesis statement at this point. The significance of research just convinces the audience that your study is worth reading.
Finally, we always recommend seeking help from your research advisor whenever you’re struggling with ideas. For a more visual idea of how to discuss the significance of your research, we suggest checking out this video .
1. Do I need to do my research before discussing its significance?
No, you’re discussing the significance of your research before you conduct it. However, you should be knowledgeable about your topic and the related literature.
2. Is the significance of research the same as its implications?
No, the research implications are potential questions from your study that justify further exploration, which comes after conducting the research.
3. Discussing the significance of research seems overwhelming. Where should I start?
We recommend the problem statement as a starting point, which reveals clues to the potential outcome of your research.
4. How can I get feedback on my discussion of the significance of my research?
Our proofreading experts can help. They’ll check your writing for grammar, punctuation errors, spelling, and concision. Submit a 500-word document for free today!
Share this article:
Post A New Comment
Got content that needs a quick turnaround? Let us polish your work. Explore our editorial business services.
5-minute read
Free Email Newsletter Template
Promoting a brand means sharing valuable insights to connect more deeply with your audience, and...
How to Write a Nonprofit Grant Proposal
If you’re seeking funding to support your charitable endeavors as a nonprofit organization, you’ll need...
9-minute read
How to Use Infographics to Boost Your Presentation
Is your content getting noticed? Capturing and maintaining an audience’s attention is a challenge when...
8-minute read
Why Interactive PDFs Are Better for Engagement
Are you looking to enhance engagement and captivate your audience through your professional documents? Interactive...
7-minute read
Seven Key Strategies for Voice Search Optimization
Voice search optimization is rapidly shaping the digital landscape, requiring content professionals to adapt their...
4-minute read
Five Creative Ways to Showcase Your Digital Portfolio
Are you a creative freelancer looking to make a lasting impression on potential clients or...

Make sure your writing is the best it can be with our expert English proofreading and editing.
- Translation
Writing the Significance of a Study
By charlesworth author services.
- Charlesworth Author Services
- 20 July, 2022
The significance of a study is its importance . It refers to the contribution(s) to and impact of the study on a research field. The significance also signals who benefits from the research findings and how.
Purpose of writing the significance of a study
A study’s significance should spark the interest of the reader. Researchers will be able to appreciate your work better when they understand the relevance and its (potential) impact. Peer reviewers also assess the significance of the work, which will influence the decision made (acceptance/rejection) on the manuscript.
Sections in which the significance of the study is written
Introduction.
In the Introduction of your paper, the significance appears where you talk about the potential importance and impact of the study. It should flow naturally from the problem , aims and objectives, and rationale .
The significance is described in more detail in the concluding paragraph(s) of the Discussion or the dedicated Conclusions section. Here, you put the findings into perspective and outline the contributions of the findings in terms of implications and applications.
The significance may or may not appear in the abstract . When it does, it is written in the concluding lines of the abstract.
Significance vs. other introductory elements of your paper
In the Introduction…
- The problem statement outlines the concern that needs to be addressed.
- The research aim describes the purpose of the study.
- The objectives indicate how that aim will be achieved.
- The rationale explains why you are performing the study.
- The significance tells the reader how the findings affect the topic/broad field. In other words, the significance is about how much the findings matter.
How to write the significance of the study
A good significance statement may be written in different ways. The approach to writing it also depends on the study area. In the arts and humanities , the significance statement might be longer and more descriptive. In applied sciences , it might be more direct.
a. Suggested sequence for writing the significance statement
- Think of the gaps your study is setting out to address.
- Look at your research from general and specific angles in terms of its (potential) contribution .
- Once you have these points ready, start writing them, connecting them to your study as a whole.
b. Some ways to begin your statement(s) of significance
Here are some opening lines to build on:
- The particular significance of this study lies in the…
- We argue that this study moves the field forward because…
- This study makes some important contributions to…
- Our findings deepen the current understanding about…
c. Don’ts of writing a significance statement
- Don’t make it too long .
- Don’t repeat any information that has been presented in other sections.
- Don’t overstate or exaggerat e the importance; it should match your actual findings.
Example of significance of a study
Note the significance statements highlighted in the following fictional study.
Significance in the Introduction
The effects of Miyawaki forests on local biodiversity in urban housing complexes remain poorly understood. No formal studies on negative impacts on insect activity, populations or diversity have been undertaken thus far. In this study, we compared the effects that Miyawaki forests in urban dwellings have on local pollinator activity. The findings of this study will help improve the design of this afforestation technique in a way that balances local fauna, particularly pollinators, which are highly sensitive to microclimatic changes.
Significance in the Conclusion
[…] The findings provide valuable insights for guiding and informing Miyawaki afforestation in urban dwellings. We demonstrate that urban planning and landscaping policies need to consider potential declines.
A study’s significance usually appears at the end of the Introduction and in the Conclusion to describe the importance of the research findings. A strong and clear significance statement will pique the interest of readers, as well as that of relevant stakeholders.
Maximise your publication success with Charlesworth Author Services.
Charlesworth Author Services, a trusted brand supporting the world’s leading academic publishers, institutions and authors since 1928.
To know more about our services, visit: Our Services
Share with your colleagues
Scientific Editing Services
Sign up – stay updated.
We use cookies to offer you a personalized experience. By continuing to use this website, you consent to the use of cookies in accordance with our Cookie Policy.
- Affiliate Program

- UNITED STATES
- 台灣 (TAIWAN)
- TÜRKIYE (TURKEY)
- Academic Editing Services
- - Research Paper
- - Journal Manuscript
- - Dissertation
- - College & University Assignments
- Admissions Editing Services
- - Application Essay
- - Personal Statement
- - Recommendation Letter
- - Cover Letter
- - CV/Resume
- Business Editing Services
- - Business Documents
- - Report & Brochure
- - Website & Blog
- Writer Editing Services
- - Script & Screenplay
- Our Editors
- Client Reviews
- Editing & Proofreading Prices
- Wordvice Points
- Partner Discount
- Plagiarism Checker
- APA Citation Generator
- MLA Citation Generator
- Chicago Citation Generator
- Vancouver Citation Generator
- - APA Style
- - MLA Style
- - Chicago Style
- - Vancouver Style
- Writing & Editing Guide
- Academic Resources
- Admissions Resources
How to Write the Rationale of the Study in Research (Examples)
What is the Rationale of the Study?
The rationale of the study is the justification for taking on a given study. It explains the reason the study was conducted or should be conducted. This means the study rationale should explain to the reader or examiner why the study is/was necessary. It is also sometimes called the “purpose” or “justification” of a study. While this is not difficult to grasp in itself, you might wonder how the rationale of the study is different from your research question or from the statement of the problem of your study, and how it fits into the rest of your thesis or research paper.
The rationale of the study links the background of the study to your specific research question and justifies the need for the latter on the basis of the former. In brief, you first provide and discuss existing data on the topic, and then you tell the reader, based on the background evidence you just presented, where you identified gaps or issues and why you think it is important to address those. The problem statement, lastly, is the formulation of the specific research question you choose to investigate, following logically from your rationale, and the approach you are planning to use to do that.
Table of Contents:
How to write a rationale for a research paper , how do you justify the need for a research study.
- Study Rationale Example: Where Does It Go In Your Paper?
The basis for writing a research rationale is preliminary data or a clear description of an observation. If you are doing basic/theoretical research, then a literature review will help you identify gaps in current knowledge. In applied/practical research, you base your rationale on an existing issue with a certain process (e.g., vaccine proof registration) or practice (e.g., patient treatment) that is well documented and needs to be addressed. By presenting the reader with earlier evidence or observations, you can (and have to) convince them that you are not just repeating what other people have already done or said and that your ideas are not coming out of thin air.
Once you have explained where you are coming from, you should justify the need for doing additional research–this is essentially the rationale of your study. Finally, when you have convinced the reader of the purpose of your work, you can end your introduction section with the statement of the problem of your research that contains clear aims and objectives and also briefly describes (and justifies) your methodological approach.
When is the Rationale for Research Written?
The author can present the study rationale both before and after the research is conducted.
- Before conducting research : The study rationale is a central component of the research proposal . It represents the plan of your work, constructed before the study is actually executed.
- Once research has been conducted : After the study is completed, the rationale is presented in a research article or PhD dissertation to explain why you focused on this specific research question. When writing the study rationale for this purpose, the author should link the rationale of the research to the aims and outcomes of the study.
What to Include in the Study Rationale
Although every study rationale is different and discusses different specific elements of a study’s method or approach, there are some elements that should be included to write a good rationale. Make sure to touch on the following:
- A summary of conclusions from your review of the relevant literature
- What is currently unknown (gaps in knowledge)
- Inconclusive or contested results from previous studies on the same or similar topic
- The necessity to improve or build on previous research, such as to improve methodology or utilize newer techniques and/or technologies
There are different types of limitations that you can use to justify the need for your study. In applied/practical research, the justification for investigating something is always that an existing process/practice has a problem or is not satisfactory. Let’s say, for example, that people in a certain country/city/community commonly complain about hospital care on weekends (not enough staff, not enough attention, no decisions being made), but you looked into it and realized that nobody ever investigated whether these perceived problems are actually based on objective shortages/non-availabilities of care or whether the lower numbers of patients who are treated during weekends are commensurate with the provided services.
In this case, “lack of data” is your justification for digging deeper into the problem. Or, if it is obvious that there is a shortage of staff and provided services on weekends, you could decide to investigate which of the usual procedures are skipped during weekends as a result and what the negative consequences are.
In basic/theoretical research, lack of knowledge is of course a common and accepted justification for additional research—but make sure that it is not your only motivation. “Nobody has ever done this” is only a convincing reason for a study if you explain to the reader why you think we should know more about this specific phenomenon. If there is earlier research but you think it has limitations, then those can usually be classified into “methodological”, “contextual”, and “conceptual” limitations. To identify such limitations, you can ask specific questions and let those questions guide you when you explain to the reader why your study was necessary:
Methodological limitations
- Did earlier studies try but failed to measure/identify a specific phenomenon?
- Was earlier research based on incorrect conceptualizations of variables?
- Were earlier studies based on questionable operationalizations of key concepts?
- Did earlier studies use questionable or inappropriate research designs?
Contextual limitations
- Have recent changes in the studied problem made previous studies irrelevant?
- Are you studying a new/particular context that previous findings do not apply to?
Conceptual limitations
- Do previous findings only make sense within a specific framework or ideology?
Study Rationale Examples
Let’s look at an example from one of our earlier articles on the statement of the problem to clarify how your rationale fits into your introduction section. This is a very short introduction for a practical research study on the challenges of online learning. Your introduction might be much longer (especially the context/background section), and this example does not contain any sources (which you will have to provide for all claims you make and all earlier studies you cite)—but please pay attention to how the background presentation , rationale, and problem statement blend into each other in a logical way so that the reader can follow and has no reason to question your motivation or the foundation of your research.
Background presentation
Since the beginning of the Covid pandemic, most educational institutions around the world have transitioned to a fully online study model, at least during peak times of infections and social distancing measures. This transition has not been easy and even two years into the pandemic, problems with online teaching and studying persist (reference needed) .
While the increasing gap between those with access to technology and equipment and those without access has been determined to be one of the main challenges (reference needed) , others claim that online learning offers more opportunities for many students by breaking down barriers of location and distance (reference needed) .
Rationale of the study
Since teachers and students cannot wait for circumstances to go back to normal, the measures that schools and universities have implemented during the last two years, their advantages and disadvantages, and the impact of those measures on students’ progress, satisfaction, and well-being need to be understood so that improvements can be made and demographics that have been left behind can receive the support they need as soon as possible.
Statement of the problem
To identify what changes in the learning environment were considered the most challenging and how those changes relate to a variety of student outcome measures, we conducted surveys and interviews among teachers and students at ten institutions of higher education in four different major cities, two in the US (New York and Chicago), one in South Korea (Seoul), and one in the UK (London). Responses were analyzed with a focus on different student demographics and how they might have been affected differently by the current situation.
How long is a study rationale?
In a research article bound for journal publication, your rationale should not be longer than a few sentences (no longer than one brief paragraph). A dissertation or thesis usually allows for a longer description; depending on the length and nature of your document, this could be up to a couple of paragraphs in length. A completely novel or unconventional approach might warrant a longer and more detailed justification than an approach that slightly deviates from well-established methods and approaches.
Consider Using Professional Academic Editing Services
Now that you know how to write the rationale of the study for a research proposal or paper, you should make use of Wordvice AI’s free AI Grammar Checker , or receive professional academic proofreading services from Wordvice, including research paper editing services and manuscript editing services to polish your submitted research documents.
You can also find many more articles, for example on writing the other parts of your research paper , on choosing a title , or on making sure you understand and adhere to the author instructions before you submit to a journal, on the Wordvice academic resources pages.
Home » Education » Significance of the Study – Ultimate Writing Guide with Example
Significance of the Study – Ultimate Writing Guide with Example
Zack Saigin
- August 29, 2023

The significance of the study in research pertains to the potential significance, relevance, or influence of the research results. It elucidates the ways in which the research contributes to the current knowledge base, addresses existing gaps, or provides new insights within a specific field of study. Whether you are composing a research paper or a thesis, a section known as the Significance of the Study ensures that your readers comprehend the impact of your work. Familiarize yourself with the process of effectively writing this crucial component of your research paper or thesis by following our comprehensive steps, guidelines, and examples.
What is Significance of the Study?
The significance of the study should capture the reader’s attention. When researchers comprehend the relevance and potential impact of the work, they can better appreciate it. Reviewers who assess the significance of the study also influence the decision to accept or reject the manuscript.
The Significance of the Study serves the purpose of providing you with an opportunity to elucidate to your readers how your research will contribute to the existing literature in your field. This is where you explain the reasons behind conducting your research and its importance to the community, individuals, and different institutions.
Clarifying the Relevance
Writing the significance of a study serves the fundamental purpose of effectively conveying the importance and value of the research being undertaken. Researchers must provide an overview of the study’s background and context, shedding light on the specific gap or problem they aim to address. Through this process, they not only establish the necessary context for their work but also lay a strong foundation upon which the rest of the study can be developed.
Guiding the Research Process
The significance of the study in research example acts as a guiding compass for researchers throughout their journey. It assists in refining research questions, structuring methodologies, and making informed decisions regarding data collection and analysis. When the purpose of the study is well-defined, researchers can navigate the complexities of the research process with better clarity and direction.
Justifying Resource Allocation
In the academic realm, finite resources such as time, funding, and expertise are available. Writing the significance of a study is a means to justify the allocation of these valuable resources. By showcasing the potential contributions and impacts of the research, researchers can demonstrate why their work deserves support and investment.
Bridging the Gap
In academia, there are limited resources like time, funding, and expertise. Articulating the significance of the study serves to validate the distribution of these precious resources. By highlighting the potential benefits and effects of the research, scholars can show why their work merits backing and investment.
Types of Significance of the Study
The significance of the study encompasses several aspects that can take different shapes, each contributing to the overall value and relevance of research. In this article, we will delve into the different forms of significance that a study can possess, illuminating the diverse ways in which research can have an impact on academia, society, and more.
Theoretical Significance
At the core of many studies lies theoretical significance. This kind of significance emerges when a study contributes to the advancement of theoretical frameworks, models, or paradigms within a specific discipline. By questioning existing theories, proposing new ones, or refining existing concepts, researchers enrich the intellectual landscape and shape the future discussions in their field.
Practical Significance
Practical significance arises when the findings of a study have direct applications in real-world contexts. Whether it involves providing insights that inform policymaking, enhancing clinical practices in healthcare, or optimizing business strategies, research with practical significance directly affects how we live, work, and make decisions across various domains.
Social Significance
Certain studies hold social significance as they address issues that deeply resonate with society. Research exploring topics such as inequality, discrimination, environmental sustainability, or mental health can draw attention to crucial societal challenges. By shedding light on these issues, researchers contribute to raising awareness, fostering empathy, and inspiring collective action.
How to Write Significance of the Study?
Significance of the Study” section in a research paper, thesis, or dissertation:
Background:
Start by providing some background information about your study. This can include a brief introduction to your subject area, the current state of research in that field, and the specific problem or question that your study focuses on.
Identify the Gap:
Demonstrate the existence of a gap in the existing literature or knowledge that requires attention, and explain how your study fills that gap. The gap may be a lack of significance of research on a specific topic, inconsistent results from previous studies, or a new problem that hasn’t been investigated yet.
State the Purpose of Your Study:
Clearly state the main objective of your research. You can frame the significance of the study as a solution to the problem or gap that you identified earlier.
Explain the Significance:
Now, describe the potential impact of your study. You can highlight how your research contributes to the existing knowledge, addresses a significance of research gap, provides a new or improved solution to a problem, influences policies or practices, or leads to advancements in a specific field or industry. It’s important to make these claims realistically, considering the scope and limitations of your study.
Identify Beneficiaries:
Identify who will benefit from your study. This could include other researchers, practitioners in your field, policy-makers, communities, businesses, or others. Explain how your findings could be used and by whom.
Future Implications:
Let’s explore the implications of your study for future research. This may involve unanswered questions, newly raised inquiries, or potential methodologies that can be suggested based on your study.
Significance of the Study Example
For instance, consider the significance of a study presented in the following fictional example:
Significance in the Introduction
Our understanding of the impact of Miyawaki forests on local biodiversity in urban housing complexes is limited. To date, no formal investigations have been conducted to examine their negative effects on insect activity, populations, or diversity. In our study, we compared the influence of Miyawaki forests on local pollinator activity within urban dwellings. The results of this significance of research can enhance the development of this afforestation technique, ensuring a harmonious coexistence with local fauna, especially pollinators, which are highly susceptible to microclimatic changes.
Significance in the Conclusion
The findings from our study offer valuable insights that can guide and inform the implementation of Miyawaki afforestation in urban dwellings. We have demonstrated the need for urban planning and landscaping policies to consider potential declines in order to mitigate any adverse effects.
You May Also Like
© 2023 by Infomatly.com
What Makes the Significance of the Study Plausible?
- January 2022
- In book: Fundamentals of research in Humanities, Social Sciences and Science Education: A practical step-by-step approach to a successful research journey (pp.30-36)
- Publisher: Van Schaik Publishers

- Fort Hare University

- University of South Africa
Discover the world's research
- 25+ million members
- 160+ million publication pages
- 2.3+ billion citations
- . . . Types Of Research Paradigms
- . . . Types
- . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Introduction
- . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Site
- . . Approaches
- . . . . . . . . . . Meanings
- . . . . . . . . . . Critical
- . . . Importance Of Research Ethics
- . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . References
- Recruit researchers
- Join for free
- Login Email Tip: Most researchers use their institutional email address as their ResearchGate login Password Forgot password? Keep me logged in Log in or Continue with Google Welcome back! Please log in. Email · Hint Tip: Most researchers use their institutional email address as their ResearchGate login Password Forgot password? Keep me logged in Log in or Continue with Google No account? Sign up
Back to blog home
7 steps to accurately test statistical significance, the statsig team, how can you ensure your insights are reliable and not just random chance.
This is where the concept of statistical significance comes into play, helping you make confident choices based on solid data.
Statistical significance is a mathematical method that measures the reliability of your analytical results. It helps you determine if the relationships or patterns you observe in your data are genuine or merely coincidental. By testing for statistical significance, you can distinguish meaningful signals from random noise, ensuring your data-backed decisions are built on a strong foundation.
Understanding statistical significance
Statistical significance is a crucial concept in data analysis that helps you assess the reliability and validity of your findings. It's a measure of how likely it is that the observed results in your data are due to a real effect or relationship, rather than just random chance. In other words, statistical significance helps you determine if the patterns or differences you see in your data are meaningful and not just flukes.
When you're analyzing data, you're often looking for patterns, trends, or relationships between variables. However, it's important to recognize that some of these observations could be due to random variation rather than a genuine effect. This is where testing for statistical significance comes in. By calculating the probability that your results could have occurred by chance alone, you can gauge the reliability of your findings.
Statistical significance plays a vital role in helping businesses make confident, data-driven decisions. By ensuring that the insights you glean from your data are statistically significant, you can:
Avoid making decisions based on false positives or random noise
Identify genuine patterns and relationships that can inform strategic choices
Allocate resources and investments towards initiatives with proven impact
Minimize the risk of costly mistakes or missed opportunities
When you're testing for statistical significance, you're essentially asking, "If there was no real effect or relationship, how likely is it that we would have observed these results by chance?" The lower this probability (known as the p-value), the more confident you can be that your findings are meaningful and not just random occurrences.
Formulating hypotheses and choosing significance levels
Hypotheses are essential for statistical testing. The null hypothesis assumes no significant difference or effect, while the alternative hypothesis contradicts the null. For example, when testing if a new feature increases user engagement, the null hypothesis would be that engagement remains unchanged.
Significance levels, denoted by α, represent the probability of rejecting a true null hypothesis (Type I error). Common choices are 0.01 and 0.05, meaning a 1% or 5% chance of incorrectly rejecting the null. A lower α reduces false positives but may miss true effects.
Choosing the right significance level depends on the consequences of Type I and Type II errors. If false positives are more costly than false negatives, a lower α is appropriate. Sample size also matters; larger samples can detect smaller effects at the same α.
To test statistical significance effectively:
Clearly define your null and alternative hypotheses based on the question you're investigating
Select an α that balances the risks of Type I and Type II errors for your specific context
Ensure your sample size is adequate to detect meaningful differences at your chosen α
Running tests at multiple significance levels can provide a more nuanced understanding of your results. While 0.05 is a common default, consider reporting results at 0.01 and 0.10 as well. This helps distinguish highly significant findings from marginally significant ones.
Multiple testing correction is crucial when testing many hypotheses simultaneously. Methods like Bonferroni correction and false discovery rate control help maintain the overall Type I error rate. Without adjustment, the chance of false positives increases rapidly with the number of tests.
By carefully formulating hypotheses and selecting appropriate significance levels, you can draw reliable conclusions from your data. Understanding how to test statistical significance empowers you to make data-driven decisions with confidence.
Collecting and analyzing data
Gathering high-quality data is crucial for accurate statistical analysis. Data collection methods include surveys, experiments, observations, and secondary data sources. Ensure data is representative of the population and minimize bias.
Statistical tests help determine if observed differences are statistically significant. T-tests compare means between two groups, while ANOVA compares means across multiple groups. Chi-square tests assess relationships between categorical variables, and Z-tests compare a sample mean to a population mean.
Sample size directly impacts the power of statistical tests. Larger samples increase the likelihood of detecting significant differences. Determine the appropriate sample size based on the desired level of significance and effect size.
When testing statistical significance , it's essential to select the appropriate test based on your data and research question. Consider factors such as data type, distribution, and independence of observations. Clearly define your null and alternative hypotheses before conducting the analysis.
Interpreting results requires understanding the p-value and significance level. A p-value below the chosen significance level (e.g., 0.05) indicates a statistically significant result. However, statistical significance doesn't always imply practical significance; consider the magnitude of the effect and its real-world implications.
Data visualization is a powerful tool for communicating statistical findings. Use graphs, charts, and tables to present results clearly and concisely. Highlight key takeaways and provide context for your audience.
Interpreting p-values and making decisions
P-values are calculated by comparing observed data to a null hypothesis using statistical tests. The p-value represents the probability of observing results as extreme as those measured, assuming the null hypothesis is true. A low p-value indicates the observed results are unlikely due to random chance alone.
To interpret p-values, compare them to a predetermined significance level (α) , typically 0.05. If the p-value is less than or equal to α, reject the null hypothesis and consider the results statistically significant. This suggests the observed effect is likely real and not just due to random chance.
However, it's crucial to avoid common misconceptions when interpreting p-values:
A p-value does not indicate the probability that the null hypothesis is true or false. It only measures the probability of observing the data if the null hypothesis were true.
A statistically significant result (p ≤ α) does not guarantee the effect is practically meaningful or important. Always consider the effect size and real-world implications alongside statistical significance.
Failing to reject the null hypothesis (p > α) does not prove the null hypothesis is true. It only suggests insufficient evidence to reject it based on the observed data.
When testing statistical significance, it's essential to:
Clearly define the null and alternative hypotheses before collecting data.
Choose an appropriate statistical test based on the data type and distribution.
Interpret p-values in the context of the study design, sample size, and potential confounding factors.
Use multiple testing correction methods (e.g., Bonferroni, Benjamini-Hochberg) when conducting numerous hypothesis tests to control the false discovery rate.
By understanding how to calculate and interpret p-values correctly, you can make informed decisions based on statistical evidence. This helps you distinguish genuine effects from random noise in your data, enabling you to confidently implement changes that drive meaningful improvements in your product or business . While statistical significance is crucial, it's not the only factor to consider. Practical relevance is equally important; a statistically significant result may not always translate to meaningful real-world impact. Assess the magnitude of the effect alongside its statistical significance.
When conducting multiple comparisons, the likelihood of false positives increases. To mitigate this, apply multiple comparison corrections such as the Bonferroni correction or the false discovery rate (FDR) method. These adjustments help maintain the overall significance level and reduce the risk of false positives.
P-hacking , the practice of manipulating data or analysis methods to achieve statistical significance, undermines the integrity of your results. To avoid p-hacking, preregister your hypotheses, specify your analysis plan in advance, and report all conducted analyses transparently. Adhering to these practices ensures the credibility of your findings.
Sample size plays a critical role in detecting statistically significant differences. When testing for statistical significance, ensure your sample size is adequate to detect meaningful effects. Conducting power analysis can help determine the appropriate sample size for your study.
Confounding variables can distort the relationship between your variables of interest, leading to misleading conclusions. When designing your study and analyzing data, identify and control for potential confounders. Randomization and stratification techniques can help minimize the impact of confounding variables.
Interpreting statistical significance requires caution. A statistically significant result does not necessarily imply causation; it only indicates an association. To establish causality, consider the study design, temporal relationship, and potential alternative explanations. Exercise caution when making causal claims based solely on statistical significance .
Request a demo
Statsig for startups.
Statsig offers a generous program for early-stage startups who are scaling fast and need a sophisticated experimentation platform.
Build fast?
Try statsig today.
Recent Posts
Introducing ai prompt experiments on statsig.
Statsig's AI Prompt Experiments allow you to run experiments for AI-powered products and gain real-time insights into what's working and what's not.
Data-driven development: A simple guide (for noobs!)
Master data-driven product development with Statsig. Simplify experimentation, make informed decisions, and accelerate your product's growth—all without complex coding.
Why you should "accept" the null hypothesis when hypothesis testing
Debunk the myth that you can never accept the null hypothesis and learn when you should by exploring the key differences between Fisher’s and Neyman-Pearson’s frameworks.
How much does a feature flag platform cost?
Use our customizable, detailed cost comparison tool and flexible pricing assumptions to find out which platform reigns supreme.
Optimizing config propagation time with target apps
Our recent optimizations to target apps significantly reduce config propagation latency, ensuring performance and stability for large-scale environments using Statsig.
Hackathon projects from Q3 2024: A top-secret sneak peek
Statsig Hackathons fuel innovation, from fixing bugs to creating new features. Check out the projects from our latest hackathon, including tons of cool AI tools!
Redirect Notice
Biosketch format pages, instructions, and samples.
A biographical sketch (also referred to as biosketch) documents an individual's qualifications and experience for a specific role in a project. NIH requires submission of a biosketch for each proposed senior/key personnel and other significant contributor on a grant application. Some funding opportunities or programs may also request biosketches for additional personnel (e.g., Participating Faculty Biosketch attachment for institutional training awards). Applicants and recipients are required to submit biosketches
- in competing applications for all types of grant programs,
- in progress reports when new senior/key personnel or other significant contributors are identified, and
- to support prior approval requests for changes in senior/key personnel status and changes of recipient organization.
NIH staff and peer reviewers utilize the biosketch to ensure that individuals included on the applications are equipped with the skills, knowledge, and resources necessary to carry out the proposed research. NIH biosketches must conform to a specific format. Applicants and recipients can use the provided format pages to prepare their biosketch attachments or can use SciENcv , a tool used to develop and automatically format biosketches according to NIH requirements.
Biosketch (Fellowship): Biographical Sketch Format Page - FORMS-H
Biosketch (non-fellowship): biographical sketch format page - forms-h.
- How to Apply — Application Guide
- Format Attachments (fonts, margins, page limits, and more)
- Research Performance Progress Report (RPPR)
- Create your biosketch here!
- Open access
- Published: 27 September 2024
LOCC: a novel visualization and scoring of cutoffs for continuous variables with hepatocellular carcinoma prognosis as an example
- George Luo 1 ,
- Toby Chen 2 &
- John J. Letterio 3 , 4 , 5
BMC Bioinformatics volume 25 , Article number: 314 ( 2024 ) Cite this article
Metrics details
The interpretation of large datasets, such as The Cancer Genome Atlas (TCGA), for scientific and research purposes, remains challenging despite their public availability. In this study, we focused on identifying gene expression profiles most relevant to patient prognosis and aimed to develop a method and database to address this issue. To achieve this, we introduced Luo’s Optimization Categorization Curve (LOCC), an innovative tool for visualizing and scoring continuous variables against dichotomous outcomes. To demonstrate the efficacy of LOCC using real-world data, we analyzed gene expression profiles and patient data from TCGA hepatocellular carcinoma samples.
To showcase LOCC, we demonstrate an optimal cutoff for E2F1 expression in hepatocellular carcinoma, which was subsequently validated in an independent cohort. Compared to ROC curves and their AUC, LOCC offered a superior description of the predictive value of E2F1 expression across various cancer types. The LOCC score, comprised of factors representing significance, range, and impact of the biomarker, facilitated the ranking of all gene expression profiles in hepatocellular carcinoma, aiding in the evaluation and understanding of previously published prognostic gene signatures. We also demonstrate that LOCC does not have the same assumptions required of Cox proportional hazards modeling for accurate analysis. Repeated sampling demonstrated that LOCC scores outperformed ROC’s AUC in discriminating predictors from non-predictors. Additionally, gene set enrichment analysis revealed significant associations between certain genes and prognosis, such as E2F target genes and G2M checkpoint with poor prognosis, and bile acid metabolism and oxidative phosphorylation with good prognosis.
In summary, we present LOCC as a novel visualization tool for the analysis of gene expression in cancer, particularly for understanding and selecting cutoffs. Our findings suggest that LOCC scores, which effectively rank genes based on their prognostic potential, represent a more suitable approach than ROC curves and Cox proportional hazard for prognostic modeling and understanding in cancer gene expression analysis. LOCC holds promise as an invaluable tool for advancing precision medicine and furthering biomarker research. Further research regarding multivariable integration and validation will help LOCC reach its full potential and establish its utility across diverse cancer types and clinical settings.
Graphical abstract

Peer Review reports
In the field of medicine, continuous markers play a crucial role in assessing an individual’s health status, offering valuable information through a range of continuous values rather than discrete categories. These markers find extensive application in medical practice, aiding in diagnosis, prognosis, and treatment planning [ 1 ]. Examples of such markers include blood glucose levels, cholesterol levels, white blood cell count, and various biomarkers like gene expression and tumor size in cancer patients [ 1 , 2 ].
Currently, there are clinical tests that utilize gene expression for cancer prognostic interpretation and treatment planning, such as Oncotype DX and Mammaprint [ 3 , 4 ]. However, despite the excitement surrounding gene profiling for hepatocellular carcinoma, gene signatures face challenges related to their functional connections and validation in independent cohorts, preventing their adoption in clinical settings [ 5 , 6 , 7 , 8 , 9 , 10 ].
Interpreting continuous markers, especially new biomarkers, can be challenging, primarily when determining the optimal threshold to divide groups [ 11 , 12 ]. In the clinical setting, knowing that variables affect outcomes is not enough as it is critical to identify relevant cut-offs that can be used for clinical decision-making. Traditional methods, like using median or quartiles, and more recent computational methods using significant p values, often result in a loss of information and fail to convey the full picture [ 1 , 11 , 13 ].
Current calculation methods, such as receiver operating characteristic (ROC) curves, have limitations in evaluating prognosis and outcome models effectively [ 14 , 15 , 16 , 17 ]. While ROC is useful for definitive diagnoses, it lacks discrimination and calibration in most prognostic studies, demanding better calculations for continuous variables and their outcomes [ 14 , 15 ]. Moreover, interpreting the meaning of the ROC area under the curve (AUC) for prognosis, where values are typically around 0.6, remains unclear.
Another commonly referenced methodology for examining the relationship between continuous variables and their associated outcomes is the Cox proportional hazard (Cox PH) model [ 18 ]. The Cox PH model is a powerful tool to analyze survival data and establish the effects of a continuous variable across an entire distribution; however, it relies on assumptions of proportionality and linearity [ 19 ]. As a result, its applicability to prognostic studies needs to be validated as it may not always hold true [ 19 , 20 ].
To address these challenges, we have developed Luo’s Optimization Categorization Curves (LOCC), an innovative tool that visualizes more information for improved cutoff selection and understanding the significance of continuous variables in relation to measured outcomes. LOCC visualizes and uses information from the entire dataset to create a graph and score to explain the prognostic cutoffs and significance of the biomarker of interest. In this paper, we present the process of generating and interpreting LOCC using practical survival curve examples with real data from The Cancer Genome Atlas [ 21 ]. We compare LOCC with ROC curve analysis and demonstrate how LOCC scores better represent prognostic value in the context of gene expression, using E2F1 as an example. Cox PH modeling was also included for comparison which highlighted the potential issues with assumptions of proportional hazard and linearity. Meanwhile, LOCC investigates the prognostic value of gene expression through categorically evaluating cut-offs within a continuous gene expression distribution with little reliance on proportionality hazards and better illustrated biomarker prognostic potential compared to Cox PH numbers. As a result, while the Cox PH model is often invaluable for survival analysis, its methodological constraints and assumption requirements make it a less direct comparison for LOCC and less accurate for cancer biomarker analysis. ROC AUC is a standard and widely accepted metric for assessing the performance of diagnostic tests and predictive models across various thresholds, making it a more relevant comparison for our LOCC approach but still we demonstrate LOCC has significant advantage in interpretability and robustness compared to ROC.
Additionally, we reanalyze several published gene signatures, illustrating how LOCC scores effectively rank each gene’s prognostic value. Moreover, we showcase how optimizing a gene signature with LOCC scores can simplify the model without compromising predictive power. The consistency and robustness of LOCC scoring are further validated through various sampling of the TCGA dataset. Finally, we explore pathways associated with prognosis in hepatocellular carcinoma.
We firmly believe that LOCC can revolutionize the analysis and understanding of continuous variables in various biological settings, providing an invaluable contribution to medical research and biomarker analysis. Through LOCC’s enhanced visualization and scoring capabilities, medical professionals can better understand biomarkers and form hypotheses to solve medical issues.
Data sources
Our study utilized two primary data sources: The Cancer Genome Atlas (TCGA) data from cBioportal.com [ 22 ] and LIRI-JP (Liver hepatocellular carcinoma – Japan) data from the International Cancer Genome Consortium (ICGC). The TCGA data was processed using z-scores of gene expression analyzed by RNA-Seq by Expected Maximization (RSEM). On the other hand, the LIRI-JP data was processed using Fragments Per Kilobase of transcript per Million mapped reads (FPKM), which was then converted to transcripts per million (TPM). Subsequently, z-scores for each sample were computed by subtracting individual expression by mean expression and dividing by the standard deviation. For TCGA data, mutation data was considered as a mutant for any non-silent mutation, which is defined as a mutation that results in different amino acids.
LOCC visualization
To generate the LOCC ranking graphs, data was processed in R. We ordered samples by expression z-scores and graphed them using ggplot. As necessary, a line representing the selected or ideal cutoff was added.
To determine the LOCC cutoff, we considered every possible categorization that would result in a distinct grouping of patient samples. We then calculated the corresponding hazard ratio (HR) and p values using the survival package in R. The HR was calculated using a Cox proportional hazard regression model, with p values evaluated via a log-rank test in R using survdiff function. The HR and p values were computed for each cutoff which resulted in at least 10% of the total population in each group. We selected the optimal cutoff with the lowest p value where each group had at least 10% of the population. We restricted the cutoffs to be at least 10% of the dataset because we wanted to reduce bias on the extremes from impacting the LOCC score. The cutoff was further verified with cutpointR [ 11 ]. For evaluation, we considered genes that were expressed in at least 20% of tumor samples.
In instances where numerous samples exhibited no gene expression, we applied a decrement of 0.1 to both the HR value and the − log ( p value) for every such sample, continuing this adjustment until the HR value reached 1 and the − log ( p value) reached 0. This adjustment served as a penalty for genes with widespread lack of expression, ensuring a more balanced assessment. We used these penalty numbers as they applied some reduction to the LOCC scores to these genes but not to the worst case scenario. The LOCC algorithm code is accessible in the data availability section.
The LOCC score is composed of three numeric components: a significance aspect, a range aspect, and an impact aspect. The three factors were chosen as they illustrate important insights about the data set. The significance aspect is denoted by − log ( p value), which illustrates the greatest statistical significance possible within the data; the range aspect is the percentage of cutoffs with a p value below 0.01 to exemplify the general strength of the relationship between the variables in the data set; the impact is the highest HR, which showcases the peak predictive power of the continuous variable. A p value of 0.01 was chosen for the range aspect as a p value less than 0.01 is a commonly used cut-off for significance in biomedical research. For significance and impact, the numbers are restricted to cutoffs such that at least 10% of the population is in each group. This restriction is to ensure that all cutoffs continue to include a substantial proportion of the population and to minimize the possibility of extremes and outliers portrayed by a nominal proportion of the sample, as these outliers are not representative of the population and can substantially skew HR p and p l values. Through extensive observation of the data, we identified that 10% is the optimal restriction to ensure statistical robustness while also minimizing biases by including a substantial proportion of patients.
The LOCC incorporates significance, range, and impact and is calculated by multiplying these three components together, represented by the equation:
where p l is the highest value of − log ( p value), R s is the percentage of cutoffs that have a highly significant p value ( p < 0.01), and HR p is the HR at the most significant point. The resulting LOCC score allows for ordering of variables from lowest to highest or vice versa, ensuring the most significant HR is above one, or its reciprocal if the most significant HR is below one.
For segregating poor prognostic markers from good prognostic markers, we assigned negative scores to all poor prognostic markers when ranking all gene expressions by LOCC score. Thus, the most positive LOCC scores were linked with good prognosis, while the most negative scores corresponded to poor prognosis. Genes expressed in at least 20% of tumor samples were evaluated. In the absence of gene expression in any tumor samples, we followed the same incremental decrease approach as mentioned in the LOCC visualization section.
LOCC cutoff estimated p value and q value
Traditional methods of estimating and interpreting p values would not work well for significance values since they are the lowest p value from a range of p values that are related to each other. To estimate a p value and q value for the most significant cutoffs for all the gene expression profiles, we applied Monte Carlo methods to estimate significance values, p l , from randomly generated data. The randomly generated data used the TCGA LIHC data but instead of being ordered by gene expression values, it was ordered through a random number generator in R. Thus, although the data had the same samples and survival times, the order was random which made it comparable to the existing results. We then calculated the LOCC scores for this randomly generated data and recorded it. We did this 10,000 times to have a large enough dataset to compare to the existing results.
Using these 10,000 random simulations, we can estimate the p value according to the empirical p values calculations [ 23 ]. Afterward, we used the empirical p values generated to calculate q values using the qvalue package in R. These q values are False Discover Rate (FDR) adjusted p values.
Cox PH modeling
We employed Cox proportional hazards (Cox PH) regression modeling to assess the association between gene expression levels and overall survival in patients. The analysis was performed using the R programming language with the survival package.
We constructed Cox PH models for each gene by treating the gene expression level as the predictor variable.
The primary Cox PH model assumed a linear relationship between gene expression and the hazard of death. To test for non-linearity, we also fit a secondary Cox PH model that included a quadratic term for gene expression. We then performed a likelihood ratio test to compare the linear and quadratic models, calculating a p values to evaluate the evidence for non-linearity.
To verify the proportional hazards assumption, we conducted a Schoenfeld residuals test using the cox.zph function, which provides a p values indicating whether the proportional hazards assumption holds for each gene. If the p value from this test was less than 0.05, it suggested a violation of the proportional hazards assumption.
For each gene, we stored the following statistics in a global summary table: the regression coefficient, the exponentiated coefficient, the standard error of the coefficient, the z-value, the p value for the gene’s effect on survival, the p value from the non-linearity test, and the p value from the proportional hazards test. The analysis was performed for each gene across the entire dataset, resulting in a comprehensive evaluation of the potential prognostic significance of gene expression levels with respect to overall survival.
Upon processing the patient and tumor data, Receiver Operating Characteristic (ROC) curves were constructed using the ROCR package in R. The Area Under the Curve (AUC) was estimated using ROCR, which took into account overall survival, excluding patients with incomplete survival or expression data. For comparative purposes, a red line representing sensitivity = (1− specificity) was included. To align with LOCC score ranking, we associated the highest AUC with good prognosis and the lowest AUC with poor prognosis. However, individual gene expression profiles or gene sets were required to have an AUC above 0.5. We evaluated genes that were expressed in at least 20% of tumor samples.
RISK gene signature
We applied the risk score analysis for hepatocellular carcinoma from a previous study [ 24 ] using the same gene expression and weight coefficients. Cox regression analysis was performed with the Cox proportional hazard package in R, and ROC analysis was conducted using the ROCR package. The proportionality assumption was verified using the function cox.zph. We selected patients who survived at least one month for ROC calculations. We used the Akaike information criterion (AIC) to assess the relative quality of models during Cox regression modeling, calculating AIC using extractAIC. In particular, for the original RISK gene signature and the 8-gene RISK gene signature, we used cox regression proportional hazard modeling of patient survival with gene expression profiles of the original RISK or the 8-gene RISK. We then used extractAIC to find the AIC of this modeling. This AIC is the relative quality of the model with a lower number being higher quality relative to the number of variables. Gene expression correlations were derived from cBioportal.com [ 22 ].
2-Fold cross validation of TCGA dataset
For the twofold cross validation, the TCGA hepatocellular data set was randomly split into two random halves; one half was used to calculate the relevant cut-offs, whereas the other half was used for validation. The procedure is as follows: first, we randomly sampled half of the TCGA hepatocellular carcinoma data. With this half, we processed the data and calculated LOCC scores, cutoffs, and ROC c-statistics. The ROC c-statistic is equivalent to the ROC AUC. We then examined the generalizability of our calculations by examining its validity within the other half of the dataset. This twofold cross validation procedure was replicated 100 times for each gene under evaluation. The cutoff with the lowest p value was chosen and tested in the validation set. The validation p value was recorded and deemed significant if it fell below 0.05.
Gene set enrichment analysis (GSEA)
After we ranked all genes by their LOCC score in hepatocellular carcinoma, we analyzed these genes and LOCC scores using pre-ranked GSEA to understand what pathways are associated with prognosis. Utilizing pre-ranked GSEA, we investigated hallmark gene sets and ranked them by their false discovery rate (FDR). FDR is a proven method to identify and minimize false positives; compared to its alternatives, FDR has been shown to be a more powerful and consistent approach, and its applicability to computational biology and genomics has been illustrated in prior research [ 25 ]. Additionally, the normalized enrichment score (NES) and p values were recorded.
To evaluate gene sets without overlapping genes, we identified and removed all overlapping genes between the gene sets. Then we used a custom gene set in pre-ranked GSEA to evaluate the FDR, NES, and p values . This will reduce the chance that a gene set is significant only due to other genes from another significant gene set.
We also evaluated gene sets of randomly selected genes to ensure p values followed a uniformed distribution. We generated 100 gene sets of 200 randomly selected genes from the TCGA expression list. We used GSEA preranked with LOCC scores to evaluate the p values from these gene sets. We compiled the p values and graphed them in Microsoft Excel and used a line of best fit to evaluate uniformity.
LOCC demonstrates E2F1 expression is associated with a poor prognosis in hepatocellular carcinoma
To demonstrate the utility of LOCC, we used TCGA data to showcase how LOCC can help analyze the role of transcription factor E2F1 in liver hepatocellular carcinoma (TCGA LIHC). E2F1 is an important transcription factor that has roles in cell cycle, DNA repair, and even apoptosis [ 26 , 27 ]. E2F1 can bind p53 to induce apoptosis and can also be inhibited by the retinoblastoma protein (Rb) to arrest the cell at the G1/S checkpoint [ 26 , 27 ]. As such, E2F1 is an important target in cancer where it is often overexpressed.
LOCC offers a comprehensive visualization of multiple parameters for continuous variables (refer to Supplemental Fig. S1 for detailed LOCC labeling). The initial graph, the LOCC ranking, presents the values of the continuous variable versus the ranking of all samples. In our instance, we plotted E2F1 z-score expression in the tumor samples on the y-axis, and the sample rankings on the x-axis (Fig. 1 A). We employed the z-score because various datasets adopt different standardization methods for RNA-Seq data, hence, we used a normalization method to approximate the distribution. We also highlighted gene mutations in different colors to observe if they affect gene expression.
LOCC demonstrates E2F1 is associated with a poor prognosis in TCGA hepatocellular carcinoma A The z-score expression of E2F1 from TCGA hepatocellular carcinoma patient samples was ordered in descending order and plotted against the ranking of the samples. Samples with mutations of E2F1 that modify the characteristics of their corresponding amino acids (non-silent mutations) are colored in orange while samples with wildtype E2F1 are colored turquoise. B A black line depicting the hazard ratio is plotted for every cutoff for E2F1 expression. A red horizontal line is placed at HR = 1.0. C A yellow line depicting the − log ( p value) is added to the graph to display the significance of each cutoff. The red horizontal line is also aligned with p = 0.01 while the green horizontal line is aligned with p = 0.05. The cutoff with the lowest p value is selected to be the ideal cutoff, indicated by the arrow which corresponds to a z-score of − 0.305. D A Kaplan–Meier overall survival curve is plotted at the ideal cutoff to separate patients into high or low E2F1 . E A data table of the details of the groups is shown. Patients’ survival times are expressed in months. P values are calculated using log-rank test. HRs are calculated using Cox proportional hazard regression. F A ROC is plotted for E2F1, and the AUC was calculated
The LOCC cutoff selection graph, which plots the hazard ratio at every single cutoff (Fig. 1 B), reveals that the hazard ratio (black line) is nearly always above 1 (red line), suggesting that higher E2F1 expression consistently correlates with a worse prognosis in LIHC.
The critical inquiry is whether E2F1 expression is truly significant at any point. To address this, we supplemented a second line, defined by its own y-axis scale, − log ( p value), on the hazard ratio graph (Fig. 1 C). When the yellow line (representing − log ( p value)) lies above the green line, it is statistically significant ( p < 0.05), and above the red line is very significant ( p < 0.01). Hence, the ideal cutoff should be within the top 50% to 75% of E2F1 expression. Using the lowest p value approach, a cutoff at 61.6% is calculated to produce the lowest p value. The appropriateness of this cut-off can be confirmed via visual inspection. Simultaneously, the precise E2F1 expression (z-score = − 0.305) at this cutoff can be visualized using the first graph shown in Fig. 1 A. We also generated a Kaplan–Meier curve at that cutoff to evaluate its appropriateness and calculate median survival (Fig. 1 D, E ). Previous literature corroborates the prognostic marker role of E2F1 gene expression in hepatocellular carcinoma using a median expression cutoff [ 28 ].
We have also developed a LOCC score to help judge significance and overall impact of any single or group of predictors. Three factors are multiplied together to get the LOCC score: the significance, the range, and the impact. The significance value is calculated by taking the − log (lowest p value). The range is the percentage of LOCC significance line (yellow line) that is above the red line ( p < 0.01). Finally, the impact is the HR at the most significant cutoff (if HR is below 1, use the reciprocal). We also limit group sizes for significance and impact to be at least 10% of the population to minimize extreme effects from small samples. Thus, the higher the LOCC score, the more critical and predictive the expression is for prognosis. In our evaluation of E2F1, the LOCC score was calculated to be 2.58 (Fig. 1 C), signifying there is some prognostic value in this dataset. In the following sections, the scale of the LOCC score will be established.
We generated a ROC curve using E2F1 expression and survival status (Fig. 1 F). With an area under the curve (AUC) of 0.578, this implies a heightened risk of death with increasing E2F1 expression. Interestingly, we observed that the red line for LOCC (HR = 1) and ROC (True Positive Rate = False Positive Rate) are derived from the same underlying equation (Supplemental Fig. S2 A–E). If the HR line remains above the red line throughout the graph, then the ROC curve will also be above the red line of random classifier. Thus, the relative positions (above/below) of the black and red lines should be similar in both LOCC and ROC curves.
Validation of E2F1 as prognostic biomarker in the LIRI-JP hepatocellular dataset A The z-score expression of E2F1 from LIRI-JP hepatocellular carcinoma patient samples was ordered in descending order and plotted against the ranking of the samples. A horizontal line is graphed at -0.305 to separate patients into high and low E2F1 groups using the TCGA dataset cutoff. B The LOCC cutoff selection was graphed for LIRI-JP samples. The cutoff from TCGA data was used to separate patients into high and low E2F1 groups. C A Kaplan–Meier overall survival curve is plotted at the validation cutoff to separate patients into high or low E2F1 . D A data table of the details of the groups is shown. Patients’ survival times are expressed in days. P values are calculated using log-rank test. HRs are calculated using cox proportional hazard regression. E A ROC is plotted for E2F1 and the AUC was calculated
Validation of E2F1 as prognostic biomarker in the LIRI-JP hepatocellular dataset
Following the identification and establishment of a significant cutoff in the TCGA data, validation in another hepatocellular carcinoma cohort is necessary. To achieve this, we utilized data from the International Cancer Genome Consortium (ICGC), which includes a large Japanese cohort of liver hepatocellular carcinoma (LIRI-JP). The LIRI-JP data provides normalized read counts, represented as Fragments Per Kilobase of transcript per Million mapped reads (FPKM), which differ from the RNA-Seq by expectation maximization (RSEM) used by TCGA. While it is feasible to reprocess both dataset’s raw data with the same programs to achieve a consistent expression format, this would be a highly demanding process. Therefore, we opted to use the normalized z-score to approximate the cutoff, despite its imperfections, which should give us a relative comparison of expression between patients.
Taking the E2F1 cutoff of -0.305 z-score established in the TCGA training, we applied this to the validation cohort (Fig. 2 A). Employing LOCC, we evaluated the significance and appropriateness of this cutoff (Fig. 2 B). Despite the TCGA cutoff not being the lowest p value of this dataset, the cutoff remains highly significant, indicating the cutoff is still appropriate. We then generated a Kaplan–Meier plot using the TCGA cutoff, to compare the survival curves of the two groups (Fig. 2 C). Given the significant survival differences observed in both TCGA and LIRI-JP cohorts using the same cutoff (Fig. 2 D), it’s reasonable to propose that E2F1 is a prognostic biomarker for LIHC, associated with poor prognosis. The LOCC score, calculated to be 18.9 (Fig. 2 B), implies strong prognostic potential of E2F1 in this dataset.
The ROC curve indicates an AUC of 0.614, suggesting increased death risk associated with higher E2F1 expression. However, determining the significance of this predictor or the optimal, significant cutoffs is challenging through the ROC curve alone. Therefore, the utility of ROC in evaluating E2F1 expression as a prognostic biomarker is limited compared to LOCC.
Comparison of ROC and LOCC in evaluating prognostic biomarkers
The use of ROC curves and their corresponding Area Under the Curve (AUC) is ubiquitous in biomarker studies and prognostic modeling [ 14 , 15 ]. However, the application and interpretation of ROC graphs and the c-statistic (or AUC) for prognostic purposes raise questions. In diagnostics, the c-statistic is akin to the probability that a randomly selected subject who experienced the event will have a higher test score than a subject who did not [ 29 ]. For prognosis, it represents the likelihood that a patient who succumbed to the disease had a higher test score than a patient who survived. Although appropriate for diagnosis, the interpretation of c-statistics is challenging for prognosis, as the binary classification of patients as alive or dead is oversimplistic for overall survival and unsuitable for censored and time-dependent data. Even the time-dependent ROC, designed to address this issue, still struggles with censoring [ 30 ].
LOCC, on the other hand, utilizes a cox regression model and examines hazard ratios between groups. Patients are classified into two categories based on the biomarker, enabling us to evaluate the hazard ratio and associated p values. Similar to ROC, LOCC is applied across all potential cutoffs and the resulting hazard ratios and p values are plotted to identify the optimal cutoff. This process aids not only in the selection of the cutoff but also enables us to assess changes in the hazard ratio across the biomarker’s range.
To compare LOCC’s and ROC’s ability to evaluate prognostic biomarkers in various cancers, we investigate E2F1 cut-offs within LIHC, Sarcoma (SARC), pancreatic ductal adenocarcinoma (PAAD), kidney renal papillary cancer (KIRP), and lower grade glioma (LGG). Different cancer types should presume different cut-offs since their gene expression and prognostic distributions differ; what is considered “high” or “low” expression for each cancer type is expected to vary based on intrinsic differences between the cancers.
The TCGA SARC dataset [ 31 ] presents an ROC curve for E2F1 expression with an AUC of 0.576 (Fig. 3 A). While the AUC for E2F1 expression in LIHC appears similar, the LOCC graph reveals notable differences, with a LOCC score for sarcoma E2F1 of only 0.11 (Fig. 3 B), much lower than the 2.58 for E2F1 in LIHC. The main difference lies in the p values and significant ranges, whereas the hazard ratios (HR) were comparable. Once the most significant cutoff was chosen, a Kaplan–Meier curve was plotted (Fig. 3 C). Despite the low LOCC score, the prognosis value of E2F1 in sarcoma is less robust than E2F1 in hepatocellular carcinoma, in stark contrast to the similar ROC c-statistics.
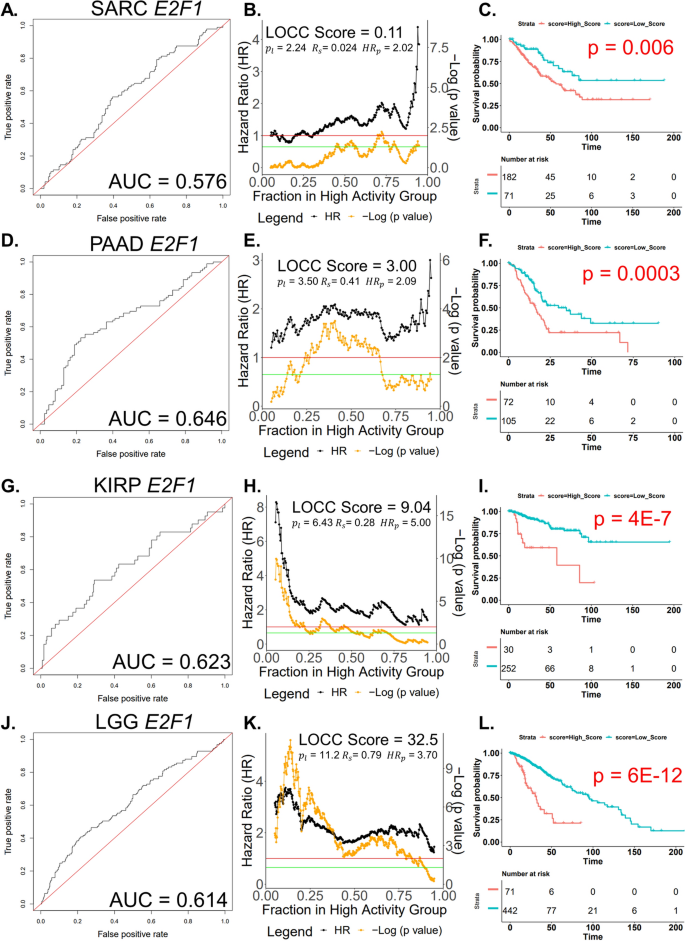
Comparison of ROC and LOCC in evaluating prognostic biomarkers A A ROC curve is plotted and the AUC was calculated for TCGA SARC E2F1 . B LOCC was plotted and scored for SARC E2F1. C The most significant cutoff was selected and a Kaplan–Meier plot was graphed to illustrate the best stratification according to E2F1 in SARC. D A ROC curve is plotted and the AUC was calculated for TCGA PAAD E2F1 . E LOCC was plotted and scored for PAAD E2F1. F The most significant cutoff was selected and a Kaplan–Meier plot was graphed to illustrate the best stratification according to E2F1 in PAAD. G A ROC curve is plotted and the AUC was calculated for TCGA KIRP E2F1 . H LOCC was plotted and scored for KIRP E2F1. I The most significant cutoff was selected and a Kaplan–Meier plot was graphed to illustrate the best stratification according to E2F1 in KIRP. J A ROC curve is plotted and the AUC was calculated for TCGA LGG E2F1 . K LOCC was plotted and scored for LGG E2F1. L The most significant cutoff was selected and a Kaplan–Meier plot was graphed to illustrate the best stratification according to E2F1 in LGG. Abbreviations and symbols: TCGA – The Cancer Genome Atlas, SARC – Sarcoma, PAAD – Pancreatic ductal adenocarcinoma, KIRP – Kidney renal papillary cell carcinoma, LGG – low grade glioma, p l is − log ( p value) at most significant cutoff, R s is percentage of cutoffs that have a highly significant p value ( p < 0.01), HR p is the HR at the most significant cutoff
In the TCGA PAAD dataset [ 32 ], we plotted the ROC curve for E2F1 , yielding an AUC of 0.646 (Fig. 3 D). With a comparably high AUC and a LOCC score of 3.00 (Fig. 3 E), both indices increased from the SARC data. However, the HR for both cancers at the most significant cutoffs was approximately 2.0, with the difference in p values across the LOCCs leading to discrepancies in scores. Using the most significant cutoff, a Kaplan–Meier plot showed significant stratification using E2F1 expression (Fig. 3 F).
In the TCGA KIRP dataset [ 33 ], the ROC curve for E2F1 revealed an AUC of 0.623 (Fig. 3 G). While the KIRP ROC curve appears similar to the PAAD curve at first glance, LOCC provides a contrasting view, identifying a high-risk group among the top 20% of E2F1 expression, while cutoffs near the median were marginally significant (Fig. 3 H). With a LOCC score of 9.04, this indicates a high prognostic potential for E2F1 in this dataset, if the correct cutoff is employed. The Kaplan–Meier curve confirms that a small proportion of KIRP patients exhibit significantly higher risk with increased E2F1 expression (F i g. 3 I).
Lastly, the TCGA LGG data [ 34 ], was evaluated with the ROC curve for E2F1 , resulting in an AUC of 0.614 (Fig. 3 J). Despite a lower AUC compared to PAAD and KIRP, the LGG LOCC score of 32.5 was significantly higher, demonstrating that most cutoffs were highly significant (Fig. 3 K). This is evident in the Kaplan–Meier curve, which identifies two distinct patient groups based on E2F1 expression (Fig. 3 L). Consequently, E2F1 expression is a significant prognostic predictor in LGG, a finding that has prompted further research into targeting this pathway [ 35 ].
While cancer types vary, ROC AUC values remain relatively similar, whereas LOCC values vary. These variabilities in LOCC correctly exemplify differences in the gene expression distributions between these cancer types. ROC AUC values seem to fluctuate minimally and somewhat randomly with no clearly defined meaning within its fluctuations, whereas LOCC scores vary based on prognostic potential. Notably, E2F1 HR values were all greater than 1 across the entire distribution of all 5 cancer types meaning that higher E2F1 is associated with worse prognosis at all points though of different statistical significance and hazard ratios. As a result, LOCC not only correctly identifies that E2F1 is associated with poor prognosis in all of these cancers but it also provides additional insight into the gene expression distribution and prognostic power through the absolute value of its score.
LOCC score helps rank prognostic importance of predictors
First, we applied LOCC analysis and scoring to all gene expression profiles in TCGA LIHC (Supplemental Table 1 ). Furthermore, we demonstrate that the top prognostic cut-offs identified by LOCC are statistically significant even after adjusting for multiple testing through Monte Carlo methods of simulation of random permutations of the dataset to estimate empirical p values and q values (Supplemental Table 1 and Supplemental Table 2 ).
Next, we demonstrate the Cox PH analysis in cancer biomarker analysis has a few critical flaws which limit its appropriateness in ranking predictors. We performed Cox PH analysis for all gene expression of LIHC (Supplemental Table 3 ). Some of the top ranked genes by Cox PH p value include GAGE family genes which are aberrant expressed genes in cancers [ 36 ]. However, only a minority of tumor samples have expression of GAGE genes and many other aberrant expressed genes and this is not clear on the Cox PH analysis summary (Supplemental Fig. S3 A, B). LOCC analysis also rank many of these genes near the top of the list but not nearly as high due the limitation that it is only expressed in a minority of samples (Supplemental Table 1 ). Furthermore, LOCC analysis clearly show that these genes have duplicate gene express levels, a sign of non-expression.
The larger issue with Cox PH analysis is that it has two major assumptions which can lead to skewed results if they are violated [ 20 , 37 ]. In the situation of cancer biomarkers, this is a significant issue as 52 of the top 100 significant genes by Cox PH p value for LIHC have violation of at least one assumption of proportional hazard or linearity (Supplemental Table 3 ). To demonstrate how this can affect accurate portrayal of the prognostic value of predictors, we compare two gene expression, POLR2H and TBP , which have similar Cox PH p values of 0.049 but differing LOCC scores of 0.0097 and 1.1, respectively.
For POLR2H , both assumptions of proportional hazard and linearity are not being violated ( p > 0.05) and thus the Cox PH p value is appropriate (Supplemental Fig. S3 C, D). However, using LOCC, we can see that this biomarker has little very significant cutoffs and a small portion in the significant range (Supplemental Fig. S3 E). As such, its LOCC score is very low at 0.0097 suggesting this gene has little prognostic potential.
For TBP , both assumptions of proportional hazard and linearity are violated ( p < 0.05) in this dataset which suggest the Cox PH p value may not be accurate (Supplemental Fig. S3 F, G). Violation of the proportional hazard means that the relationship between the covariate and the hazard rate is not constant over time, which can lead to incorrect or misleading estimates of hazard ratios in the Cox PH model. Violation of the linearity assumption indicates that the relationship between a continuous covariate and the log hazard is non-linear which can lead to biased estimates of hazard ratios and incorrect conclusions about the effect of the covariate. However, it is not clear if this will lead to overestimation or underestimation of the p value but simply that Cox PH analysis may be inaccurate. Using LOCC, we can visualize what is happening with TBP that is causing problems with Cox PH analysis. Violation of the proportional hazard leads to varying hazard ratios over time which may be seen in highly fluctuating hazard ratios on the LOCC graph seen in Supplemental Fig. S3 H. Similarly, violation of linearity is visualized in LOCC when hazard ratios show significant inflection points. In this scenario, the LOCC is better in assessing the prognostic potential of the predictor since certain portions of the gene expression range are useful for prognostic evaluation while other portions are not. Cox PH uses the entire range which is not appropriate in this scenario when only a certain portion is prognostic significant. Biologically, this can occur for many reasons such as needing a threshold for pathway activation or where overexpression of a gene has diminishing effects. LOCC visualization show TBP is significantly associated with poor expression at high expression but that low expression is not associated with good prognosis. Cox PH modeling only show that the assumptions are violated and thus fails to provide accurate commentary regarding the prognostic potential of this biomarker.
LOCC scores also have better prognostic interpretability and understanding compared to ROC score. With the LOCC score, a marker scoring zero signifies a lack of prognostic value; an AUC of 0.5 in the ROC score system also implies no predictive value. Yet, the threshold for the ROC AUC is nebulous as almost all genes register an AUC above or below 0.5.
We utilize TCGA hepatocellular carcinoma data to compare the prognostic value of LOCC scores vs AUC scores and confirm their validity by comparing identified prognostic biomarkers with previous literature. The following thresholds were established for this analysis; LOCC scores > 0.1, > 1, and > 8. An LOCC score above 0.1 means there exists some highly significant prognostic cutoff; an LOCC score greater than 1 means that the corresponding cancer biomarker has a sizable range of highly significant cutoffs (> 10%) along with other appropriate significance values and hazard ratios for biomarker consideration; an LOCC score above 8 represents the top 1% of LOCC scores in this TCGA LIHC dataset and these biomarkers have the highest potential prognostic value.
We evaluated 18,789 genes within TCGA hepatocellular carcinoma. 5398 (28.7%) showed LOCC scores above 0.1 or below − 0.1, 2455 (13.1%) had LOCC scores above 1 or below − 1, and 214 (1.1%) had LOCC scores above 8 or below − 8 (Fig. 4 A, Supplemental Table 1 , Table 1 ). With these percentages, we identified ROC AUC cutoffs that pinpointed a similar proportion of genes. An ROC AUC above 0.54 or below 0.46 encompassed 6301 (33.5%) genes; an ROC AUC above 0.56 or below 0.44 included 2731 (14.5%) genes; an ROC AUC above 0.60 or below 0.40 contained 203 (1.1%) genes (Fig. 4 B, Supplemental Table 1 , and Table 1 ). Absolute AUC cutoffs of 0.54, 0.56, and 0.60 corresponded most closely to absolute LOCC scores of 0.1, 1, and 8; these thresholds were utilized to compare the predictive power of LOCC scores vs AUC scores and assess the potential for enrichment.
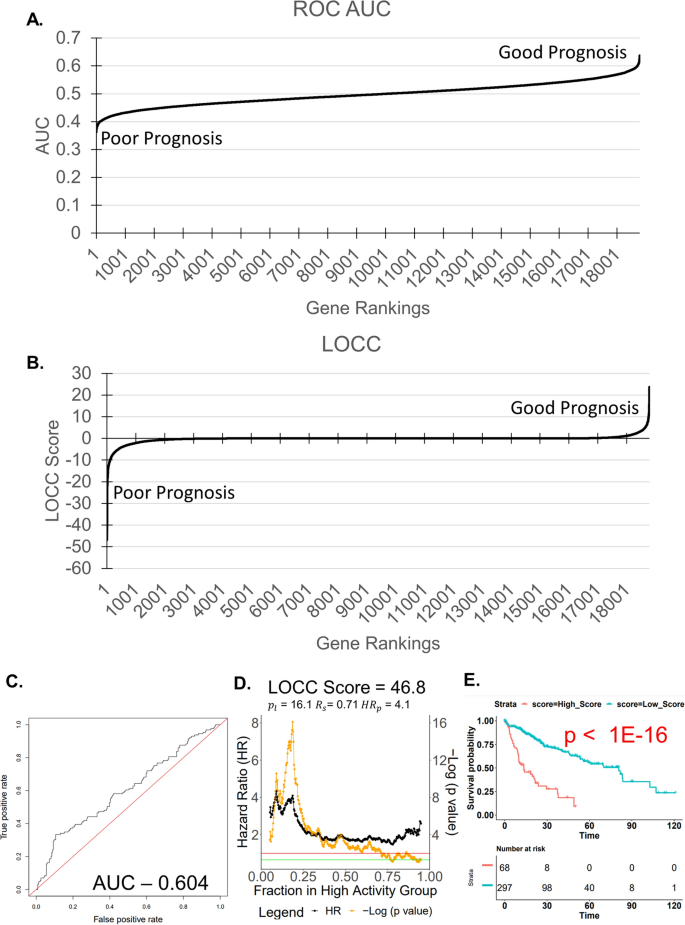
LOCC score helps rank prognostic importance of predictors A Genes were evaluated and ranked by ROC AUC. B Genes were evaluated and ranked by LOCC score. C The ROC curve is shown for TCGA hepatocellular carcinoma KIF20A expression and the AUC is calculated. D The LOCC cutoff selection is graphed for TCGA hepatocellular carcinoma KIF20A expression. E A Kaplan–Meier curve is plotted for KIF20A at the most significant cutoff. Patients’ survival times are expressed in days. P values are calculated using log-rank test. HR are calculated using Cox proportional hazard regression
Notably, LOCC has a clear correlation between absolute value and predictive power and a clear score cut-off for delineating prognostic potential. A LOCC score of zero suggests no cutoffs can achieve highly significant prognostic importance ( p < 0.01). A LOCC score of 1 may indicate potential prognostic value, as this score corresponds to a substantial proportion (~ 0.1–0.2) of highly significant cutoffs (Supplemental Table 1 ). In contrast, the literature lacks a clearly defined set AUC for prognosis, which is instinctively challenging to establish as evidenced in the example above.
Four recent publications describe prognostic gene signatures using the TCGA hepatocellular carcinoma dataset [ 24 , 38 , 39 , 40 ]. These works identify 34 distinct genes potentially useful as hepatocellular carcinoma predictors. However, the predictor overlap between the four publications is minimal, with only two genes (SPP1 and LECT2) found common in two of the datasets.
We believe this discrepancy stems from the use of Least Absolute Shrinkage Selector Operator (LASSO) regression, which selects good predictors without offering a holistic view of the disease landscape. The variability among different publications indicates a need for a better approach to understand the full picture of hepatocellular carcinoma predictors. To help better understand the full picture of hepatocellular carcinoma predictors and their importance, we identified these 34 genes by their ranking by LOCC score and ROC AUC to investigate any trends.
We summarize the categorization of scores of all ranked genes and the 34 different genes in Table 1 and the full data regarding individual predictor genes in Supplemental Table 4 . The trend is that 34 predictor genes are clustered at the top of LOCC scores and ROC AUC. However, it appears that the LOCC score better explains the selection of gene predictors. Using a cutoff of absolute LOCC score greater than 1, there were 2455 genes (13.1% of all genes) fitting that condition and contain 28 of the 34 predictor genes (Table 1 ). Meanwhile, using a cutoff of absolute ROC AUC greater than 0.56, there were 2731 genes (14.5% of all genes) fitting that condition and also contain 28 of the 34 predictor genes (Table 1 ). Using a cutoff of absolute LOCC score greater than 8 or absolute ROC AUC over 0.60 yields 214 genes (1.1% of total). Of the top 214 LOCC score genes, 14 of them were from the 34 gene predictors (Table 1 ). On the other hand, of the top 203 genes by ROC AUC (AUC > 0.6) there were only 9 out of the 34 predictors (Table 1 ). Finally, the average percentile of gene predictors in each gene signature was lower using LOCC score ranking compared to ROC AUC ranking (Supplemental Table 4 ).
Thus, while it appears that the gene predictors were clustered near the top of both scores, the LOCC score consistently better explained the selection of the gene predictors. In particular, we compare the ROC AUC and LOCC visualization and score for the KIF20A , a predictor in one of the published gene signatures [ 24 ] (Fig. 4 C–E ). The ROC AUC of KIF20A is 0.60, which is in the top 200 but its LOCC score is 46.8, the highest of all ranked genes. From the LOCC visualization, there is one very distinct peak and there is a very significant survival difference between the groups using that cutoff (Fig. 4 D, E ).
Next, we demonstrate even among the predictors in the gene signatures, LOCC score can help select which genes are contributing significantly or not as significantly. We used LOCC scoring to rank the 12 individual genes from a previously published prognostic gene signature (RISK) in hepatocellular carcinoma [ 24 ]. We found the top eight gene expression by LOCC score ( KIF20A, TTK, TPX2, LCAT, SPP1, HMMR, CYP2C9, ANXA10 ) had a significantly higher LOCC score than the other 4 genes (LOCC Score < 0.05 and log 10 LOCC p value < 0.05, Table 2 ). ROC AUC of the 12 genes were all between 0.57 and 0.62 with 10 genes between 0.59 and 0.62 though the top 8 were significantly higher in ROC AUC than the bottom 4 genes ( p < 0.05, Table 2 )[ 24 ]. Meanwhile, cox regression analysis also found a significant difference in the − log ( p value) between the 8 genes from LOCC analysis and the other 4 genes ( p = 0.0007, Table 2 ).
We tested if this new gene signature (8-gene RISK) would be as useful as the original 12 as it contained the most significant genes according to LOCC, ROC, and cox regression. To do this, we used the same weight coefficient as the original article and used it to calculate the 8-gene RISK score [ 24 ]. Indeed, we found that the optimized gene signature had a similar p value and HR between the high and low risk groups compared to the original 12 gene signature (Supplemental Fig. S4 A–E). Furthermore, the LOCC score, AUC of the ROC curves, and cox regression analysis of the 8-gene RISK and original 12-gene RISK produced very similar numbers. (Supplemental Fig. S4 D–G). Finally, the Akaike information criterion (AIC), a number to measure relative model quality, shows that the 8-gene RISK score is a similar if not better predictor model (Supplemental Fig. S4 E). Overall, from the extensive analysis and comparisons, we believe the gene signature from the top 8 genes by LOCC score is non-inferior to the original 12 gene signature demonstrating LOCC score ability to decipher the key predictors.
While the LOCC score is correlated with the cox regression p values and ROC AUC (Supplemental Fig. S4 A, B), we believe LOCC score provides more information, particularly for prognosis. One example of the difference between the cox regression p value and LOCC score is the rankings of KIF20A , TTK , and TPX20. Although the three gene expression profiles are strongly correlated (Supplemental Fig. S4 D, E), LOCC scoring values very significant cutoffs while cox regression and ROC AUC favor having a more consistent HR (Table 2 ) . While the LOCC score does favor significant cutoffs, it is not as singular as using only the most significant cutoff as required by some established methods [ 11 , 13 ]. Instead as a hybrid score, LOCC is more holistic and can function without adjustment, though future studies will be needed to evaluate LOCC score utility in more gene expression prognosis analysis. However, we argue that LOCC score better represents the overall gene signature. This can be seen as KIF20A and TTK are ranked higher by LOCC score than TPX2 while TPX2 has higher ROC AUC and cox regression p value than both KIF20A and TTK (Table 2 ). When we compare the LOCC visualization of the original 12-gene or 8-gene RISK signature (Supplemental Fig. S3 A, B), both the HR and significance lines more closely resemble the LOCC visualization of KIF20A and TTK compared to TPX2 (Fig. 4 D, Supplemental Fig. S4 F, G). This suggests that the contribution of KIF20A and TTK are likely more significant than TPX2 in shaping the overall prognostic gene signature.
Variability and reproducibility of LOCC scores and ROC AUC
To scrutinize the consistency and reproducibility of LOCC scores and ROC AUC, we explored the gene expression of three distinct prognostic markers within a diverse sampling of the TCGA dataset. The markers selected include KIF20A , which has the highest LOCC score and is a critical predictor in the RISK score[ 24 ] (Fig. 4 C–E), E2F1, a verified prognostic indicator presented in Fig. 1 and 2 ; and MTHFR, a marker previously identified in a hepatocellular carcinoma prognostic gene set [ 38 ] (Supplemental Fig. S6 A, B). The LOCC scores and ROC AUC of KIF20A , E2F1 , and MTHFR respectively are 46.7, 2.58, 0.01 and 0.604, 0.578, 0.528 (Supplemental Table 5 ). The trends revealed by these scores suggest a coherent pattern among the three genes, a pattern that we aim to sustain and clarify in random samples from the dataset.
twofold cross-validations were utilized to examine the generalizability and variability of the data set. Upon conducting a 100 twofold cross-validation, we discerned that the general trend was maintained across all features investigated, yet the degree of variability differed by feature (Fig. 5 A–F). Significant HR, highest − log ( p value), and percent highly significant each demonstrated a preserved trend with non-overlapping interquartile ranges (Fig. 5 A–C and Table 2 ). The best cut, determined by the cutoff with the most significant p value, exhibited considerable variability for MTHFR , minimal for E2F1 , and virtually none for KIF20A (Fig. 5 D and Supplemental Table 5 ). The LOCC scores effectively differentiated between the three genes though with significant variability for KIF20A (Fig. 5 E and Table 2 ). Finally, ROC AUC exhibited considerable variability but also had considerable overlap between E2F1 and KIF20A AUC (Fig. 5 F and Table 2 ).
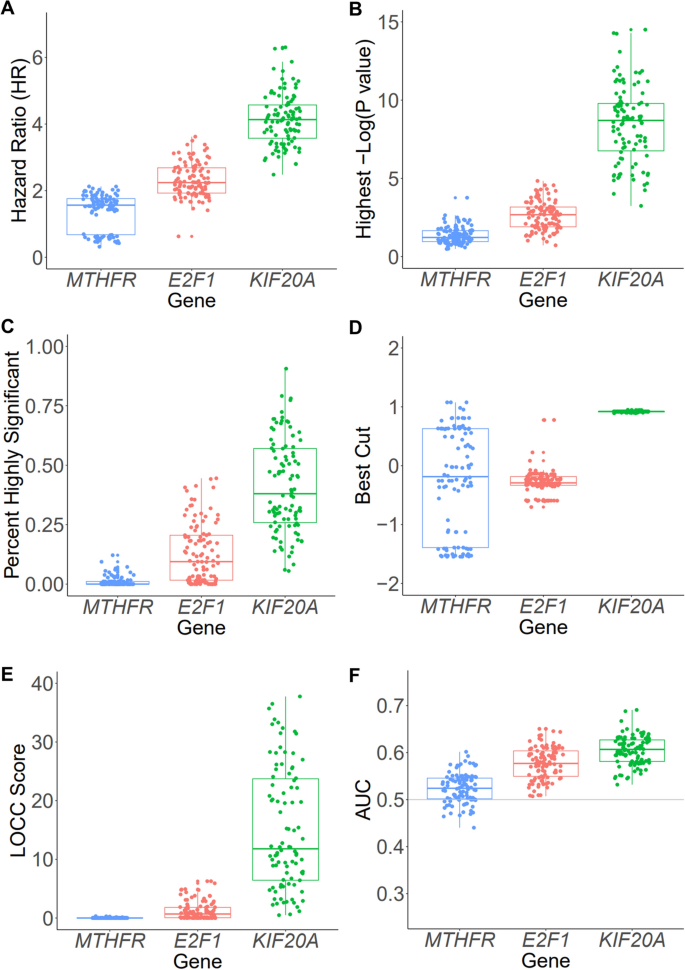
Variability and Reproducibility of LOCC Scores and ROC AUC A The 8-gene modified RISK score was ordered in descending order and plotted against the ranking of the samples. A horizontal line is graphed at 2.00 to separate patients into high and low risk groups using ideal LOCC cutoff. B The LOCC cutoff selection was graphed for the 8-gene RISK score using TCGA hepatocellular data. The most significant cutoff was chosen to separate patients into two groups. In addition, another cutoff at 1.077 was selected for dividing patients into three groups. C A Kaplan–Meier overall survival curve is plotted at the validation cutoff to separate patients into high or low risk score. D A comparison of the RISK score of 8 leading genes by LOCC score and the original 12 gene RISK score is shown. The lowest p value and hazard ratio (HR) are selected with a minimum of at least 10% of the total samples in each group. E Low risk patients are further stratified using the LOCC cutoff selector into middle and low risk. A corresponding Kaplan–Meier graph is plotted for the three different risk group. Patients’ survival times are expressed in months. P values are calculated using log-rank test. HR are calculated using Cox proportional hazard regression
Comparison of features between the full dataset and individual samples reveals several patterns (Table 2 ). The significant HR is relatively consistent between the full dataset and the samples. However, the highest − log ( P value) and percent highly significant are considerably lower in the samples, a foreseeable outcome given the dependency of p values on the number of samples. Consequently, the LOCC scores are on average lower in the samples as compared to the full dataset. The ROC AUC was similar on average between the samples and the full dataset but demonstrated substantial variability among individual samples. The optimal cutoffs were similar for the full dataset and individual samples, with the exception of MTHFR , which exhibited three distinct clusters of points, leading to the discrepancy (Fig. 5 D). Further, leveraging the twofold cross validation, we were able to investigate the “validation success rate” of our data set. The validation success rate represented the percentage of the twofold cross-validation that exhibited p < 0.05 when using the full data best cut-off. This percentage represents the probability that the calculated LOCC cutoff can be replicated or validated in miniature cohort; if the twofold cross-validation success rate is high, then the likelihood of validation using similar methods is high as the variance within the data set is low, and if the twofold cross-validation success rate is low, then the likelihood of validation using methods is low due to high variance within the data set. With this approach, we examined the validation success rate of KIF20A , E2F1 , and MTHFR . Their validation success rates were 100%, 62%, and 1%, respectively. Notably, KIF20A successfully passed every twofold cross-validation while E2F1 passed the majority, but MTHFR only managed 1% of the twofold cross-validations, suggesting its limited utility as a prognostic predictor (Supplemental Table 5 ).
To better illustrate certain samples, we chose three from MTHFR and KIF20A for further investigation using LOCC (Supplemental Fig. S6 C, D). By comparing the visualization of the full dataset MTHFR and the three samples of MTHFR , we observed both similarities and distinctions (Supplemental Fig. S6 A, C). The most dramatic changes occur at the edges, yet each sample bears resemblance to the full dataset. KIF20A , on the other hand, showcases a distinct peak at the high score group (Fig. 5 D), a feature retained in all its samples (Supplemental Fig. S6 D). High peaks, marked by the yellow significant line, imply that the cutoff is more resilient to sampling bias, offering another interpretation of p values. However, we did notice edge noise, particularly in sample 3 of KIF20A . Despite the high HR at the right edge, the yellow line does not surpass the red line. As such, the optimal cutoff remains the same as the original where the yellow line peaks around 0.20. In conclusion, we believe LOCC visualization and scoring effectively upheld their integrity during the variability and reproducibility sampling, underscoring their utility as computational tools for the analysis and comprehension of cutoffs and prognostic predictors.
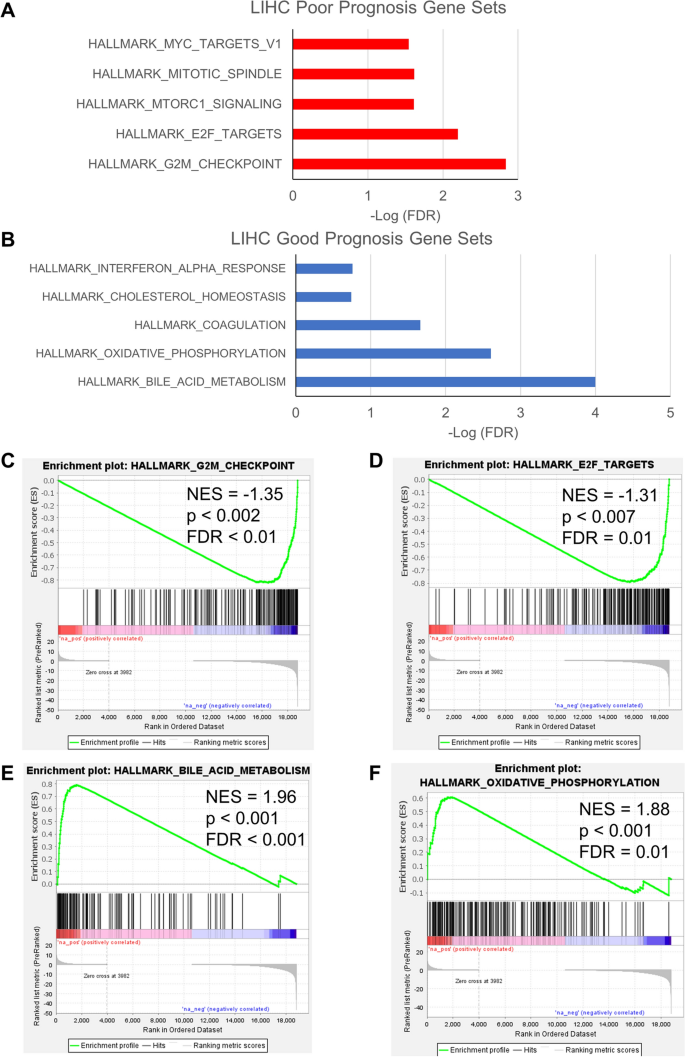
Individual gene expression and gene signatures associated with prognosis in hepatocellular carcinoma A The most significant poor prognosis gene sets from GSEA are shown. Gene sets are sorted by FDR (False Discovery Rate). B The most significant good prognosis gene sets from GSEA are shown. Gene sets are sorted by FDR (False Discovery Rate). C The enrichment plot of the hallmark G2M checkpoint is shown. D The enrichment plot of the hallmark E2F targets is shown. E The enrichment plot of the hallmark bile acid metabolism is shown. F The enrichment plot of the hallmark oxidative phosphorylation is shown
Individual gene expression and gene signatures associated with prognosis in hepatocellular carcinoma
While individual gene expression profiles are interesting biomarkers for diseases, it is of greater biological relevance to understand which pathways are of prognostic significance, which can be achieved through LOCC analysis. Leveraging LOCC scores, we can rank each gene based on its prognostic potential. Then, by utilizing GSEA, we can determine the key physiological pathways and gene sets associated with the individual genes identified by LOCC. While LOCC classifies the prognostic value of individual genes, further examining LOCC scores with GSEA allows us to discern which gene sets and pathways are crucial for prognosis. We applied this approach to hepatocellular carcinoma, scoring 18,789 genes with LOCC, out of which 10,145 genes exhibited non-zero LOCC scores. Employing these LOCC scores (Supplemental Table 1 ), we utilized GSEA pre-rank enrichment to identify their associated prognostic pathways and gene clusters. A handful of hallmark pathways emerged as significantly relevant (Fig. 6 A, B, Supplemental Table 6 ). In particular, G2M checkpoint and E2F targets correlated with poor prognosis, while bile acid metabolism and oxidative phosphorylation were linked to good prognosis (Fig. 6 C–F). These findings align with previous research [ 39 ], further confirming the validity of the LOCC scores. However, our method also flagged oxidative phosphorylation, which was likely overlooked in the prior study as they focused on contrasting gene expression patterns between two groups, rather than examining all genes in a continuous manner.
To ensure the robustness of our GSEA model, we also examined possible redundancy and overlap between gene signatures. Between bile acid metabolism and oxidative phosphorylation, we identified only 4 overlapping genes and over 100 distinct genes within each gene set. As such, its redundancy is negligible and will have minimal to no impact on GSEA. On the contrary, hallmark E2F targets and G2M checkpoints have significant overlap with 73 overlapping genes and 127 distinct genes in each gene set (Supplemental Fig. S7 A). We conducted a secondary GSEA to determine whether overlapping gene signatures would meaningfully impact our GSEA results. When overlapping genes are removed, we observe very similar NES, p values, and FDR values for each gene set (Supplemental Fig. S7 B, C), illustrating that the overlapping portion of genes does not significantly alter the overall analysis and confirming that G2M checkpoint and E2F targets are associated with poor prognosis.
We also performed validation that the p values from preranked GSEA of randomly created gene sets followed an expected uniform distribution of p values. We generated 100 gene sets of up to 200 genes per set and performed preranked GSEA using LOCC scores which did demonstrate an uniform distribution of p values (Supplemental Fig. S7 D, Supplemental Table 6 ). As such, the very low p values for the top Hallmark pathways associated with prognosis are unlikely to due to pure chance.
In this study, we introduce LOCC, an innovative visualization tool that depicts the distribution, significance, and hazard ratios associated with a continuous variable. This enables the selection of an optimized cutoff or multiple cutoffs while surveying the entire range of potential cutoffs. While this study investigates hepatocellular carcinoma, LOCC is not limited to analysis within cancer. LOCC can analyze any data set containing a continuous variable and its corresponding prognostic outcomes. In this study, we’ve demonstrated the applicability of LOCC towards elucidating the relationship between gene expression and hepatocellular carcinoma prognosis. However, we anticipate that LOCC may be applied towards a broad spectrum of carcinomas and even other diseases as a tool to examine the relationship of a continuous variable and its prognostic outcomes.
LOCC offers several advantages over prior methods, especially ROC curve and Cox PH model analysis for prognosis. By fusing the hazard ratio and p value into a single graph, the significance and impact of a variable can be comprehended simultaneously. We established that LOCC holds more informative content compared to ROC curves through direct comparison. In the context of prognosis, sensitivity, specificity, and the c-statistic are challenging to interpret, while HR and p values are straightforward. LOCC analysis is not also limited by the assumptions that are required by Cox PH to have accurate results. Thus, we propose the LOCC visualization and scoring tool as a valuable resource for the analysis of many continuous variables to outcome analyses, potentially outperforming ROC curves and Cox PH analysis [ 17 ].
We suggested the LOCC score as a representative metric for ranking variables by significance, range, and impact. Using a combination of three variables, we can generate a score to help pinpoint the most significant variables related to the outcome. This score represents our attempt to numerically encapsulate the comprehensive LOCC visualization for ease of understanding and sorting. The LOCC score outperforms ROC for prognostic studies, as the AUC has a relatively small range and large variability in sampling tests. Even though both LOCC score and ROC AUC assist in enriching prognostic gene predictors, the LOCC score demonstrated superior enrichment performance for gene predictors. A notable difference is LOCC’s incorporation of significance into its scoring, which aids in determining valuable predictors of outcome, while ROC AUC cutoffs appear arbitrary. The inclusion of significance, range, and impact into the LOCC score allows LOCC to more effectively portray the prognostic value of a biomarker; a biomarker with no prognostic value will always score a low LOCC score and vice versa. Additionally, biomarkers with an LOCC score of zero can easily be categorized as having little to no prognostic value; the ROC AUC approach does not clearly delineate what is considered a significant vs insignificant prognostic number. As noted previously ROC AUC cutoffs are arbitrary at best, and ROC AUC cutoffs are not always correlated with prognostic potential. As a result, the LOCC score better characterizes the relationship between a gene set and prognostic outcomes. All in all, LOCC is a more practical, robust approach for prognostic evaluation.
As discussed previously, the Cox PH model has also been utilized to characterize prognostic studies, but its reliance on the assumption of proportionality and linearity across an entire distribution within a data set limits its practicality and feasibility. The Cox PH model often overlooks cutoffs identified by LOCC as the Cox PH model is unable to account for non-proportional associations within datasets [ 41 ]. In many cases, particularly in complex biological prognostic studies, continuous variables hold predictive value at certain ranges but do not hold linearity and proportionality across an entire data distribution, which can cause biased estimates and misleading information when relying on the Cox PH model [ 19 , 37 ]. This can be seen in our specific examples of when assumptions of Cox PH model was violated leading to misleading p values. The LOCC model resolves this limitation by employing an alternative approach; by identifying specific cutoffs within the data set that hold significant prognostic value, we can measure the predictive power of a data set with or without an assumption of proportionality. Further, the LOCC is able to pinpoint specific cut-offs with the greatest prognostic relevance. As a result, LOCC provides a more practical realistic methodology to accurately examine the prognostic power of continuous variables and their associated outcomes.
There are still challenges in the bioinformatics field regarding the categorization of continuous variables. Within the context of cancer and gene expression, LOCC can assist in ranking variables and visualizing the relevance of the variable and outcome, but its multi-variable integration needs refinement. In our example, we utilized weighted coefficients from the original study that applied LASSO (Least Absolute Shrinkage Selector Operator) regression. Given that many models like LASSO employ single weight approaches, novel methods might be necessary for selecting and weight variables for optimal LOCC score and prognosis prediction [ 42 ]. This notion stems from the fact that LASSO regression may omit potentially active predictors, especially if predictors are correlated or if the number of predictors isn’t significantly larger than the number of samples [ 42 , 43 ]. We saw this kind of issue where several studies used LASSO regression to select prognostic gene signatures for hepatocellular carcinoma with minimal overlapping genes. Furthermore, these prognostic gene signatures often lack biological significance or understanding, leading others to ignore them. Ideally, a gene signature should be prognostic and contain elements that can be interpreted individually or as a group. In addition, we plan to integrate other aspects of cancer data such as cancer stage and age of the patient to better characterize prognostic markers. Therefore, the challenge of finding a superior method for integrating these variables remains [ 44 ].
A notable advantage of LOCC is its ability to visualize the selection process and hazard risks, comparable to showing one’s work in mathematics. This allows the audience to understand which cutoffs are selected, how they were chosen, and how the validation process succeeded or faltered. Despite many calculations being computer-based, it becomes easier to validate others’ work by checking if LOCC graphs align. Moreover, when validation doesn’t proceed as expected, it is possible to visualize what went wrong, such as the absence of a peak or a weaker than expected signal. Consequently, LOCC can supplement existing and future prognosis studies by presenting a complete overview of the predictor or biomarker, increasing the audience’s confidence.
Limitations
One main limitation of LOCC is that there is while cut-offs are ranked in order, this could be of very minor numerical difference in the continuous variable. A small change in the continuous variable could lead to large relative ranking changes and some effects on the overall LOCC graph and score. However, with large samples and internal sampling, this limitation will likely be minor. Additionally, future methods could include a noise validation test to introduce a noise multiplier to simulate noisy measurements in the real-world though additional experiments should be performed to determine how much noise to signal is present in the model.
LOCC does heavily rely on the best specific cutoff for its rankings which can be problematic. However, the LOCC score includes a range component, helping to counteract overfitting at a specific cutoff. One aspect of LOCC that is both a limitation and benefit is the dependency on the p value, meaning that large datasets can perform better with significance. However, it is also an advantage to incorporate p values to assist in our understanding and selection of prognostic predictors. Ultimately, the best way to verify a predictor is through external validation with another separate dataset.
Finally, this work is primarily focused on the univariate analysis of LOCC. We know that univariate analysis using gene expression has limitations in understanding and predicting prognosis in the big picture of cancer; however, we do demonstrate that LOCC has advantages over ROC and Cox PH in our univariate analysis, a good sign for future development of LOCC. Multivariable visualization and integration will be helpful in better understanding and prognostic modeling of cancer biomarkers but extensive work and comparisons will be required to incorporate these aspects.
LOCC serves as a powerful visualization and prognostic tool, adept at representing the extensive data of continuous variables in relation to survival or prognosis. It facilitates the selection of optimal cutoffs and provides a comprehensive analysis of the relationship of the variable with a particular outcome. The LOCC score excels in pinpointing variables that correlate with survival, and its capacity to rank predictors has proven instrumental in elucidating prognostic genes and pathways. Further, when paired with GSEA, it can be used to identify key physiological pathways which may substantiate our understanding of the biological underpinnings of prognosis and survival. Serving as a viable alternative to ROC curves and Cox PH for prognosis, LOCC brings forth superior visualization and scoring capabilities, consequently providing a deeper insight into predictors and outcomes with better reliability and clarity.
Availability of data and materials
Gene expression and mutation data was accessed from cBioportal.com for TCGA LIHC and https://dcc.icgc.org/projects/LIRI-JP for LIKI-JP. Code and instructions for LOCC on R is available at https://github.com/gluo30/locc .
Bennette C, Vickers A. Against quantiles: categorization of continuous variables in epidemiologic research, and its discontents. BMC Med Res Methodol. 2012;12:21.
Article PubMed PubMed Central Google Scholar
Eng KH, Schiller E, Morrell K. On representing the prognostic value of continuous gene expression biomarkers with the restricted mean survival curve. Oncotarget. 2015;6(34):36308–18.
Sparano JA, Gray RJ, Makower DF, Pritchard KI, Albain KS, Hayes DF, Geyer CE Jr, Dees EC, Goetz MP, Olson JA Jr, et al. Adjuvant chemotherapy guided by a 21-gene expression assay in breast cancer. N Engl J Med. 2018;379(2):111–21.
Article CAS PubMed PubMed Central Google Scholar
Cardoso F, van’t Veer LJ, Bogaerts J, Slaets L, Viale G, Delaloge S, Pierga JY, Brain E, Causeret S, DeLorenzi M, et al. 70-Gene signature as an aid to treatment decisions in early-stage breast cancer. N Engl J Med. 2016;375(8):717–29.
Article CAS PubMed Google Scholar
Iizuka N, Oka M, Yamada-Okabe H, Nishida M, Maeda Y, Mori N, Takao T, Tamesa T, Tangoku A, Tabuchi H, et al. Oligonucleotide microarray for prediction of early intrahepatic recurrence of hepatocellular carcinoma after curative resection. Lancet. 2003;361(9361):923–9.
Marquardt JU, Galle PR, Teufel A. Molecular diagnosis and therapy of hepatocellular carcinoma (HCC): an emerging field for advanced technologies. J Hepatol. 2012;56(1):267–75.
Article PubMed Google Scholar
Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T, Durnez A, Demetris AJ, Thorgeirsson SS. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40(3):667–76.
Kim K, Zakharkin SO, Allison DB. Expectations, validity, and reality in gene expression profiling. J Clin Epidemiol. 2010;63(9):950–9.
Pinyol R, Montal R, Bassaganyas L, Sia D, Takayama T, Chau GY, Mazzaferro V, Roayaie S, Lee HC, Kokudo N, et al. Molecular predictors of prevention of recurrence in HCC with sorafenib as adjuvant treatment and prognostic factors in the phase 3 STORM trial. Gut. 2019;68(6):1065–75.
Qian Y, Daza J, Itzel T, Betge J, Zhan T, Marmé F, Teufel A. Prognostic cancer gene expression signatures: current status and challenges. Cells. 2021;10(3):648.
Budczies J, Klauschen F, Sinn BV, Győrffy B, Schmitt WD, Darb-Esfahani S, Denkert C. Cutoff finder: a comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS ONE. 2012;7(12): e51862.
Altman DG, Royston P. The cost of dichotomising continuous variables. BMJ. 2006;332(7549):1080.
Nagy Á, Munkácsy G, Győrffy B. Pancancer survival analysis of cancer hallmark genes. Sci Rep. 2021;11(1):6047.
Cook NR. Statistical evaluation of prognostic versus diagnostic models: beyond the ROC curve. Clin Chem. 2008;54(1):17–23.
Zou KH, O’Malley AJ, Mauri L. Receiver-operating characteristic analysis for evaluating diagnostic tests and predictive models. Circulation. 2007;115(5):654–7.
Berrar D, Flach P. Caveats and pitfalls of ROC analysis in clinical microarray research (and how to avoid them). Brief Bioinform. 2012;13(1):83–97.
Grund B, Sabin C. Analysis of biomarker data: logs, odds ratios, and receiver operating characteristic curves. Curr Opin HIV AIDS. 2010;5(6):473–9.
Abd ElHafeez S, D’Arrigo G, Leonardis D, Fusaro M, Tripepi G, Roumeliotis S. Methods to analyze time-to-event data: the cox regression analysis. Oxid Med Cell Longev. 2021;2021:1302811.
Stensrud MJ, Hernán MA. Why test for proportional hazards? JAMA. 2020;323(14):1401–2.
Rulli E, Ghilotti F, Biagioli E, Porcu L, Marabese M, D’Incalci M, Bellocco R, Torri V. Assessment of proportional hazard assumption in aggregate data: a systematic review on statistical methodology in clinical trials using time-to-event endpoint. Br J Cancer. 2018;119(12):1456–63.
TCGA. GA Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169(7):1327–41.
Article Google Scholar
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4.
North BV, Curtis D, Sham PC. A note on the calculation of empirical P values from Monte Carlo procedures. Am J Hum Genet. 2002;71(2):439–41.
Ouyang G, Yi B, Pan G, Chen X. A robust twelve-gene signature for prognosis prediction of hepatocellular carcinoma. Cancer Cell Int. 2020;20:207.
Glickman ME, Rao SR, Schultz MR. False discovery rate control is a recommended alternative to Bonferroni-type adjustments in health studies. J Clin Epidemiol. 2014;67(8):850–7.
Kent LN, Leone G. The broken cycle: E2F dysfunction in cancer. Nat Rev Cancer. 2019;19(6):326–38.
Meng P, Ghosh R. Transcription addiction: can we garner the Yin and Yang functions of E2F1 for cancer therapy? Cell Death Dis. 2014;5(8): e1360.
Huang YL, Ning G, Chen LB, Lian YF, Gu YR, Wang JL, Chen DM, Wei H, Huang YH. Promising diagnostic and prognostic value of E2Fs in human hepatocellular carcinoma. Cancer Manag Res. 2019;11:1725–40.
Austin PC, Steyerberg EW. Interpreting the concordance statistic of a logistic regression model: relation to the variance and odds ratio of a continuous explanatory variable. BMC Med Res Methodol. 2012;12:82.
Kamarudin AN, Cox T, Kolamunnage-Dona R. Time-dependent ROC curve analysis in medical research: current methods and applications. BMC Med Res Methodol. 2017;17(1):53.
TCGA. Comprehensive and integrated genomic characterization of adult soft tissue sarcomas. Cell. 2017;171(4):950–65.
TCGA. Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell. 2017;32(2):185–203.
Linehan WM, Spellman PT, Ricketts CJ, Creighton CJ, Fei SS, Davis C, Wheeler DA, Murray BA, Schmidt L, Vocke CD, et al. Comprehensive molecular characterization of papillary renal-cell carcinoma. N Engl J Med. 2016;374(2):135–45.
Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, Cooper LA, Rheinbay E, Miller CR, Vitucci M, Morozova O, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–98.
Wang X, Wang H, Xu J, Hou X, Zhan H, Zhen Y. Double-targeting CDCA8 and E2F1 inhibits the growth and migration of malignant glioma. Cell Death Dis. 2021;12(2):146.
Gjerstorff MF, Ditzel HJ. An overview of the GAGE cancer/testis antigen family with the inclusion of newly identified members. Tissue Antigens. 2008;71(3):187–92.
Matsuo K, Purushotham S, Jiang B, Mandelbaum RS, Takiuchi T, Liu Y, Roman LD. Survival outcome prediction in cervical cancer: cox models vs deep-learning model. Am J Obstet Gynecol. 2019;220(4):381.e381-381.e314.
Liu GM, Zeng HD, Zhang CY, Xu JW. Identification of a six-gene signature predicting overall survival for hepatocellular carcinoma. Cancer Cell Int. 2019;19:138.
Zhou T, Cai Z, Ma N, Xie W, Gao C, Huang M, Bai Y, Ni Y, Tang Y. A novel ten-gene signature predicting prognosis in hepatocellular carcinoma. Front Cell Dev Biol. 2020;8:629.
Chen R, Zhao M, An Y, Liu D, Tang Q, Teng G. A prognostic gene signature for hepatocellular carcinoma. Front Oncol. 2022;12: 841530.
Bewick V, Cheek L, Ball J. Statistics review 12: survival analysis. Crit Care. 2004;8(5):389–94.
Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33(1):1–22.
Tibshirani R, Bien J, Friedman J, Hastie T, Simon N, Taylor J, Tibshirani RJ. Strong rules for discarding predictors in lasso-type problems. J R Stat Soc Ser B Stat Methodol. 2012;74(2):245–66.
McDermott JE, Wang J, Mitchell H, Webb-Robertson BJ, Hafen R, Ramey J, Rodland KD. Challenges in biomarker discovery: combining expert insights with statistical analysis of complex omics data. Expert Opin Med Diagn. 2013;7(1):37–51.
Download references
Acknowledgements
The graphical abstract was created with Biorender.
G.L is supported by NIH MSTP training grant 5T32GM007250. J.J.L. is supported by the Jane and Lee Seidman Chair in Pediatric Cancer Innovation.
Author information
Authors and affiliations.
Department of Pathology, Case Western Reserve University School of Medicine, 2103 Cornell Rd., Wolstein Research Bldg. Rm 3501, Cleveland, OH, 44106, USA
School of Medicine, University of Michigan, Ann Arbor, MI, USA
The Angie Fowler Adolescent and Young Adult Cancer Institute, University Hospitals Rainbow Babies & Children’s Hospital, Cleveland, OH, USA
John J. Letterio
The Case Comprehensive Cancer Center, Cleveland, OH, USA
Department of Pediatrics, Case Western Reserve University, Cleveland, OH, USA
You can also search for this author in PubMed Google Scholar
Contributions
George Luo: Conceptualization, Methodology, Software, Validation, Visualization, Writing—original draft, Writing—review & editing. Toby Chen: Conceptualization, Validation, Writing—review & editing. John J Letterio: Supervision, Validation, Writing—review & editing.
Corresponding author
Correspondence to George Luo .
Ethics declarations
Ethics approval and consent to participate.
Not applicable.
Consent for publication
Competing interests.
The authors declare that they have no competing interests.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary material 1., supplementary material 2., supplementary material 3., supplementary material 4., supplementary material 5., supplementary material 6., supplementary material 7., rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/ .
Reprints and permissions
About this article
Cite this article.
Luo, G., Chen, T. & Letterio, J.J. LOCC: a novel visualization and scoring of cutoffs for continuous variables with hepatocellular carcinoma prognosis as an example. BMC Bioinformatics 25 , 314 (2024). https://doi.org/10.1186/s12859-024-05932-1
Download citation
Received : 22 July 2023
Accepted : 16 September 2024
Published : 27 September 2024
DOI : https://doi.org/10.1186/s12859-024-05932-1
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Gene expression
BMC Bioinformatics
ISSN: 1471-2105
- General enquiries: [email protected]
- Open access
- Published: 14 September 2024
Extreme trophic tales: deciphering bacterial diversity and potential functions in oligotrophic and hypereutrophic lakes
- Guijuan Xie 1 , 2 ,
- Yuqing Zhang 2 , 3 ,
- Yi Gong 2 ,
- Wenlei Luo 2 , 4 &
- Xiangming Tang 2
BMC Microbiology volume 24 , Article number: 348 ( 2024 ) Cite this article
105 Accesses
1 Altmetric
Metrics details
Oligotrophy and hypereutrophy represent the two extremes of lake trophic states, and understanding the distribution of bacterial communities across these contrasting conditions is crucial for advancing aquatic microbial research. Despite the significance of these extreme trophic states, bacterial community characteristics and co-occurrence patterns in such environments have been scarcely interpreted. To bridge this knowledge gap, we collected 60 water samples from Lake Fuxian (oligotrophic) and Lake Xingyun (hypereutrophic) during different hydrological periods.
Employing 16S rRNA gene sequencing, our findings revealed distinct community structures and metabolic potentials in bacterial communities of hypereutrophic and oligotrophic lake ecosystems. The hypereutrophic ecosystem exhibited higher bacterial α- and β-diversity compared to the oligotrophic ecosystem. Actinobacteria dominated the oligotrophic Lake Fuxian, while Cyanobacteria, Proteobacteria, and Bacteroidetes were more prevalent in the hypereutrophic Lake Xingyun. Functions associated with methanol oxidation, methylotrophy, fermentation, aromatic compound degradation, nitrogen/nitrate respiration, and nitrogen/nitrate denitrification were enriched in the oligotrophic lake, underscoring the vital role of bacteria in carbon and nitrogen cycling. In contrast, functions related to ureolysis, human pathogens, animal parasites or symbionts, and phototrophy were enriched in the hypereutrophic lake, highlighting human activity-related disturbances and potential pathogenic risks. Co-occurrence network analysis unveiled a more complex and stable bacterial network in the hypereutrophic lake compared to the oligotrophic lake.
Our study provides insights into the intricate relationships between trophic states and bacterial community structure, emphasizing significant differences in diversity, community composition, and network characteristics between extreme states of oligotrophy and hypereutrophy. Additionally, it explores the nuanced responses of bacterial communities to environmental conditions in these two contrasting trophic states.
Peer Review reports
Introduction
Lakes serve as sentinels and integrators, reflecting the impacts of environmental changes on watersheds [ 1 , 2 , 3 ]. Recently, the escalation of eutrophication, driven by excessive nutrient input such as nitrogen and phosphorus, poses a grate threat to the health and stability of lake ecosystems [ 4 ]. Lakes are categorized into oligotrophic, mesotrophic, and eutrophic (mild eutrophic, moderate eutrophic, and hypereutrophic) based on biological productivity and nutritional indices [ 5 ]. The consequential effects of eutrophication on biotic communities, particularly microbial communities, in lakes are noteworthy [ 6 ].
Bacteria, integral to aquatic ecosystems, play pivotal roles in biogeochemical cycles of organic matter and nutrients [ 7 , 8 ]. Minor environmental variations in aquatic settings can impact bacterial community activity [ 9 ], underscoring their critical role in signaling environmental changes and sustaining the health and stability of aquatic ecosystems. Bacterial communities influence ecosystem functions and stability through parameters like diversity, species composition, interspecific relationships, and nutrient cycling via decomposition of organic compounds [ 6 , 10 ]. Understanding variations in bacterial community structure among lakes with different trophic states is essential in aquatic microbial ecology.
The positive correlation between bacterial abundance and lake eutrophication is well-established [ 11 , 12 ]. The nutritional status of lakes alters the taxonomic structure of bacterial communities, impacting metabolic pathways of carbon, nitrogen, sulfur, and other elements [ 13 , 14 ]. Eutrophic lakes exhibit higher richness and diversity [ 15 ] but can also experience reduced diversity due to excessive nutrients [ 16 ]. Mild eutrophic lakes, in a transition state, have the most microbial species but weak species interactions [ 6 , 17 ]. However, scant attention has been given to bacterial communities in oligotrophic and hypereutrophic lakes. Comparing bacterial community structures in these extremes could offer valuable insights for the preservation and remediation of highly nutritious lakes.
Eutrophic lakes often witness the proliferation of Cyanobacteria, including toxic Microcystis and Planktothrix [ 18 , 19 ]. Cyanobacteria, Proteobacteria and Bacteroidetes dominate in eutrophic lakes [ 20 , 21 ], while oligotrophic lakes, characterized by clear water and low nutrient content, are dominated by Actinobacteria and Proteobacteria [ 7 , 22 ]. Nevertheless, research on the seasonal and spatial changes in bacterial community structure, especially interactions among bacterial species, in extreme oligotrophic and eutrophic environments remains limited.
Species in ecosystems form intricate interactions through substance, energy, and information exchange, creating complex ecological networks [ 23 ]. Bacteria contribute to these networks, with changes in structure affecting ecosystem function and stability [ 24 , 25 ]. While higher species diversity is often associated with more complex networks [ 26 , 27 , 28 ], studies show a negative or nonlinear correlation between network complexity and biodiversity [ 29 , 30 ]. Ecosystem complexity contributes to stability, as observed in macroecology [ 23 ]. Exploring the impact of climate change on microbial networks reveals a strong correlation between network complexity and stability [ 31 ]. Yet, the complexity of bacterial species interactions and how it affects community stability between oligotrophic and hypereutrophic lakes remain unclear.
In this study, we selected two freshwater lakes characterized by distinct trophic states. Lake Fuxian, an oligotrophic deep lake, contrasts with Lake Xingyun, a hypereutrophic shallow lake. The close proximity of these lakes, coupled with their unique trophic states, renders them ideal for in-depth exploration of variations in bacterial diversity and the intricate dynamics of network complexity and stability driven by their trophic conditions. Our primary objectives include: (1) To analyze and compare the bacterial diversity and taxonomic composition between Lake Fuxian and Lake Xingyun, which have distinctly different nutrient status. We hypothesize that Lake Fuxian, being oligotrophic, will exhibit lower bacterial diversity compared to the hypereutrophic Lake Xingyun. Additionally, we expect distinct taxonomic profiles reflective of their respective nutrient conditions. (2) To assess and compare the complexity and stability of bacterial networks in both lakes. We hypothesize that the bacterial networks in Lake Xingyun will demonstrate greater complexity due to the high nutrient availability, whereas Lake Fuxian’s bacterial networks will show lower stability, characteristic of oligotrophic conditions and lower species diversity. This study provided insights into how varying nutrient conditions influence bacterial communities and their ecological interactions, offering perspectives on how freshwater ecosystems might respond to eutrophication, which are escalating global concerns.
Materials and methods
Both Lake Fuxian and Lake Xingyun are situated on the Yunnan Plateau in Southwest China (Fig. 1 a). Lake Fuxian, as a representative plateau deep-water freshwater lake, boasts a surface area of 216 km 2 and an average depth of 89.6 m. It constitutes 78% of the total water storage in Yunnan Province, presenting a clear water body deficient in nutrients (Fig. 1 b), indicative of an oligotrophic status [ 32 ].
Lake Xingyun, on the other hand, is a shallow lake with a surface area of 34.7 km 2 and an average depth of 5.3 m [ 33 ]. Its hydrological recharge depends on atmospheric precipitation and the seasonal inflow from 14 small rivers [ 34 ]. The average nutrient states of Lake Xingyun is characterized as hypereutrophic (Fig. 1 c). Since 2002, the lake has witnessed annual cyanobacterial blooms. The two lakes are interconnected by the Gehe River, spanning approximately 2.0 km. In 2008, to safeguard the water quality of Lake Fuxian, the local government initiated the “Lake Fuxian-Lake Xingyun Outflow Diversion Project” aimed at blocking the Gehe River.
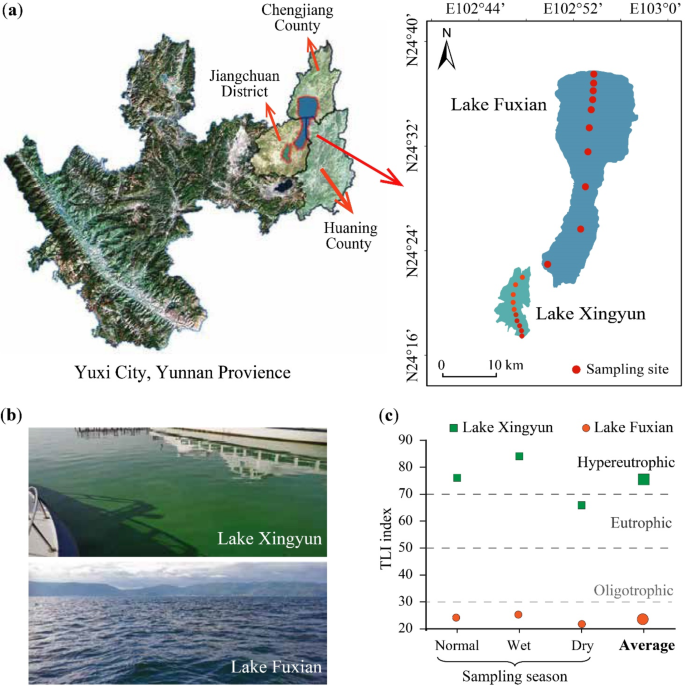
( a ) Geographical positioning of Lake Fuxian and Lake Xingyun within Yunnan Province, along with the delineation of sampling sites. ( b ) Upper: July 2020 images depicting Lake Xingyun’s water body covered with dense cyanobacterial blooms; Lower: Contrastingly, Lake Fuxian’s water body appears clear. ( c ) The comprehensive trophic level index (TLI) for Lake Fuxian and Lake Xingyun. TLI values indicate trophic states: TLI < 30 signifies oligotrophic, 30 ≤ TLI ≤ 50 denotes mesotrophic, 50 < TLI ≤ 60 implies mild eutrophic, 60 < TLI ≤ 70 indicates moderate eutrophic, and TLI > 70 represents hypereutrophic
Sample collection and environmental attribute analysis
Surface water samples of twenty liters each (top 50 cm) were collected from 10 designated sites in each lake (Fig. 1 a) during hydrological normal (April 2020), wet (July 2020), and dry (January 2021) seasons using a 5 L Schindler sampler. Transparency (SD) was measured in situ by a Secchi disk. In situ measurements of pH, water temperature (WT), dissolved oxygen (DO), and conductivity (Cond) were performed using a multiparameter water quality sonde (YSI 6600 V2, Yellow Springs Instruments Inc., USA). All samples were processed in the laboratory within 4 h.
For DNA extraction, water samples (200 ml for Lake Xingyun and 1000 ml for Lake Fuxian) were filtered through a 0.2 μm pore-size Polycarbonate membrane (Millipore Boston, MA, USA) using a vacuum pump, and the filtered materials were stored at − 80℃. The remaining water samples underwent chemical and biological analyses. Spectrophotometric methods were used to measure total nitrogen (TN), total dissolved nitrogen (TDN), ammonia nitrogen (NH 4 -N), nitrate nitrogen (NO 3 -N), total phosphorus (TP), total dissolved phosphorus (TDP), phosphate phosphorus (PO 4 -P) and chlorophyll- a (Chl- a ) content. Total suspended solids (SS) concentration was calculated by filtering 200 ~ 500 ml water samples through a pre-dried and weighed GF/C membrane, followed by drying in an oven at 105℃ for 4 h and weighing. Subsequently, the samples underwent further processing in a muffle furnace at 550℃ for 2 h. Afterward, they were cooled in a desiccator and weighed again to ascertain the concentration of inorganic suspended solids (ISS). The disparity between SS and ISS was defined as the loss on ignition (LOI) [ 35 ]. The concentration of dissolved organic carbon (DOC) was determined using a total organic carbon analyzer with the standard method (HJ/T 104–2003). Permanganate index (COD Mn ) was determined by the potassium permanganate method (GB/T 15456 − 2008).
For algal and bacterial abundance counting, 5 ml of raw water was taken into a centrifuge tube, stored at 4 °C for later algal biomass counting, and another 5 ml of raw water was mixed with 0.24 ml of formaldehyde, stored at 4℃ overnight, then transferred to -20℃ for total bacterial abundance (TB) counting. Subsequently, the samples were brought back to the laboratory and counted using a flow cytometer [ 36 ].
The trophic state assessment of the two lakes utilized the comprehensive trophic level index (TLI), a widely adopted measure for assessing the eutrophication degree of Chinese lakes. In this study, three water environmental parameters, Chl- a , TN, and TP, were employed to calculate TLI [ 37 ].
DNA extraction and sequencing
Total bacterial DNA were extracted from filter membranes using the FastDNA ® Spin Kit for soil kit (MP Biomedicals, USA) following the manufacturer’s instructions. The V3–V4 variable regions of the bacterioplankton 16S rRNA gene were PCR amplified using universal primer sets 341 F (5’-ACTCCTACGGGAGGCAGCAG-3’) and 806R (5’-GGACTACHVGGGTWTCTAAT-3’) [ 38 ]. The PCR amplification system includes a 20 ng DNA template, 0.4 µM primer, and NEB Phusion High-Fidelity PCR mix. PCR amplification program: pre-denaturation at 98 °C for 3 min; denaturation at 98 °C for 45 s, annealing at 55 °C for 45 s, extension at 72 °C for 45 s, a total of 30 cycles; final extension at 72 °C for 7 min. The amplified product was purified by Ampure XP magnetic bead method and dissolved in Elution Buffer to complete the construction of the genomic library. Finally, the DNA library was subjected to paired-end sequencing (2 × 300 bp) using the Illumina Miseq PE300 platform (Illumina, USA) of BGI Co. Ltd (Shenzhen, China).
Bioinformatic analysis of sequencing data
The raw 16S rRNA gene sequencing data underwent bioinformatic analysis using the QIIME2 Core 2023.2 distribution [ 39 ]. The process included importing, merging paired reads, splicing, and quality control of the original sequences. Sequences with lengths exceeding 550 bp or falling below 200 bp, as well as primers and other low-quality nucleotide sequences, were removed. Amplicon sequence variants (ASVs) were generated, and taxonomic classification was accomplished by comparing representative sequences against the SILVA v138 database [ 40 ]. To minimize random sequencing errors, low-abundance ASVs (< 10 sequences) were excluded from the ASV table. The raw sequencing reads have been deposited in the Genome Sequence Archive (GSA) database ( http://gsa.big.ac.cn ) under accession number CRA008654.
Bacterial diversity, taxonomic and predictive functional differences, and statistical analyses
Bacterial diversity analysis was performed using the vegan package in R (version 4.1.2). Kruskal-Wallis tests were employed to assess differences in physicochemical parameters between Lake Fuxian and Lake Xingyun, as well as across various hydrological periods. Bacterial α-diversity indices, namely the Chao1 index and Shannon index, were calculated at a uniform sequence depth (i.e., 36,377). Kruskal-Wallis tests were then applied to discern differences in bacterial α-diversity indices between the two lakes and across different sampling seasons.
For bacterial β-diversity, non-metric multidimensional scaling (NMDS) using Bray–Curtis distance among bacterial communities was performed [ 41 ]. Analysis of Similarities (ANOSIM) was employed to compare differences in bacterial community compositions (BCCs) between Lake Fuxian and Lake Xingyun, as well as among different sampling seasons. Following detrended correspondence analysis (DCA), redundancy analysis (RDA) was selected to explore associations between environmental variables and bacterial community structures. Prior to analysis, ASV data underwent Hellinger transformation. The vegan package’s forward selection method screened environmental factors with significant impacts on community composition, retaining those with a variance inflation factor (VIF) of less than 10 to mitigate collinearity issues. Significance was confirmed through a Monte Carlo test with 999 permutations. The “rdacca.hp” package was employed to evaluate the independent effect of each significant variable on the variation of BCCs [ 42 ].
To identify differentially abundant bacterial taxa between the two lakes, we employed LEfSe (linear discriminant analysis effect size) on ASVs with > 0.05% relative abundance [ 43 ]. This analysis was conducted using the online Galaxy interface ( http://huttenhower.sph.harvard.edu/galaxy ) with a Kruskal-Wallis alpha value of 0.01 and a linear discriminant analysis (LDA) threshold score of 4.0.
Functional predictive analysis was conducted using the Functional Annotation of Prokaryotic Taxa (FAPROTAX) tool on an online platform ( http://www.cloud.biomicroclass.com/ ) [ 44 ], and results were visualized using STAMP (version 2.1.3). A P -value < 0.05 was considered statistically significant.
Network construction and visualization
Molecular ecological networks were constructed using the Network Analysis Pipeline (iNAP: http://mem.rcees.ac.cn:8081/ ) [ 45 ], based on ASV data. Prior to network analysis, ASVs with a relative abundance < 0.08% were filtered out, and Spearman correlation coefficients were then calculated. The correlation matrix underwent scanning with the cutoff set from 0.01 to 1, in increments of 0.01, to determine the optimal threshold conforming to the Poisson distribution, guided by the random matrix theory (RMT) within the iNAP program. To ensure comparability between lakes, a uniform similarity threshold of 0.87 was selected for constructing co-occurrence networks.
Key network properties, including total nodes, total links, R square of power-law, average degree (avgK), average path distance (GD), and average clustering coefficient (avgCC), were calculated to describe network size. Module division was carried out using greedy modularity optimization [ 46 ]. Additionally, one hundred random networks were generated by rewiring all nodes and links corresponding to empirical networks. Parameters of both random and empirical networks were subjected to t -tests to assess the meaningfulness of network construction.
The stability of co-occurrence networks was evaluated by randomly removing 50% of the nodes from the static network, measuring the rate of reduction in network robustness [ 31 ]. Visualization and analysis of networks were performed using the interactive platform Gephi (version 0.9.2) [ 47 ], in conjunction with Cytoscape (version 3.7.1) [ 48 ] .
Characteristics of environmental parameters
The average TLI for Lake Fuxian and Lake Xingyun was 24 and 76, respectively, indicating oligotrophic and hypereutrophic conditions (Fig. 1 c). Figure 2 illustrates the average water quality parameters of both lakes. Kruskal-Wallis tests revealed significant differences in measured physicochemical (SD, pH, Cond, TN, TDN, NO 3 -N, NH 4 -N, TP, TDP, PO 4 -P, COD Mn , DOC, Chl- a , SS, and ISS) and biological parameters (abundances of algae and total bacteria) between the two lakes ( P < 0.001), except for WT and DO ( P > 0.05). The content of organic matter (OM%) in SS in Lake Fuxian was significantly higher than in Lake Xingyun ( P < 0.01).
Furthermore, environmental parameters exhibited heterogeneity during different hydrological periods (Supplementary Table S1 ). Water temperature (WT), pH, Cond, TDP, PO 4 -P, and COD Mn during the wet season were significantly higher than during the normal and dry seasons ( P < 0.05). In Lake Xingyun, TN concentrations were significantly higher in the wet season compared to the normal and dry seasons ( P < 0.05), whereas TN did not show significant variation in Lake Fuxian. The highest abundance of total bacteria (TB) in Lake Xingyun occurred in the normal season, while in Lake Fuxian, it peeked during the wet season.
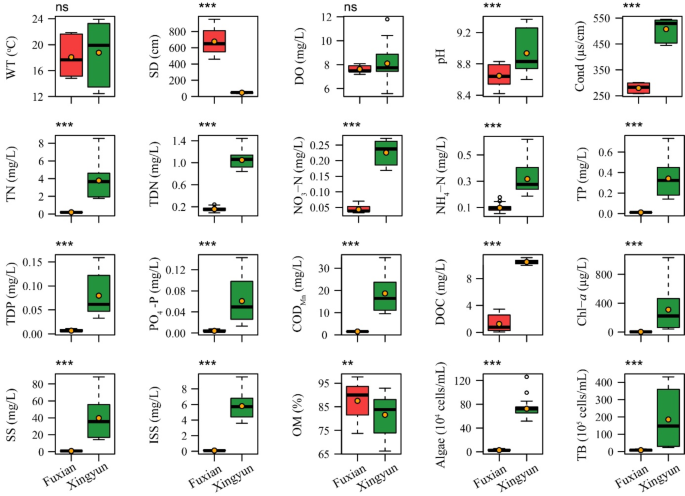
Comparison of the main environmental parameters between Lake Fuxian and Lake Xingyun. WT, water temperature; SD, Secchi disk transparency; DO, dissolved oxygen; Cond, conductivity; TN, total nitrogen; TDN, total dissolved nitrogen; NO 3 -N, nitrate nitrogen; NH 4 -N, ammonia nitrogen; TP, total phosphorus; TDP, total dissolved phosphorus; PO 4 -P, phosphate phosphorus; COD Mn , permanganate index; DOC, dissolved organic carbon; Chl- a , chlorophyll- a ; SS, suspended solids; ISS, inorganic suspended solids; OM, organic matter content; TB, total bacterial abundance. The non-parametric Kruskal-Wallis test was performed to examine differences among the lakes. At the top of each boxplot: NS indicates no significant differences ( P > 0.05); **, P < 0.01; ***, P < 0.001. In the boxplot, bold short black line and yellow dot denote the median and the mean of each parameter, respectively
Diversity of bacterial communities
Both Chao1 and Shannon indices in Lake Fuxian were significantly lower than those in Lake Xingyun (Fig. 3 ; P < 0.001). In Lake Fuxian, the Chao1 index was significantly higher in the dry and normal seasons compared to the wet season ( P < 0.001), while the Shannon index in the normal season was significantly higher than in the dry and wet seasons ( P < 0.01). In Lake Xingyun, the dynamics of the Chao1 index exhibited a trend similar to that of the oligotrophic Lake Fuxian, with the highest value occurring in the dry season. However, the Shannon index showed no significant differences across different hydrological periods ( P > 0.05).
NMDS illustrated distinct separation of BCCs based on nutrient status between lakes and among hydrological periods within each lake (Fig. 3 c). ANOSIM indicated that BCCs between the oligotrophic Lake Fuxian and hypereutrophic Lake Xingyun were significantly different (ANOSIM R = 0.99, P < 0.001). The BCCs among different hydrological periods in both lakes exhibited significant separation ( R = 0.99, P < 0.001). Additionally, in the NMDS plot, the points representing different sampling seasons in Lake Xingyun are more dispersed compared to Lake Fuxian, indicating higher β-diversity of bacterial communities in Lake Xingyun.
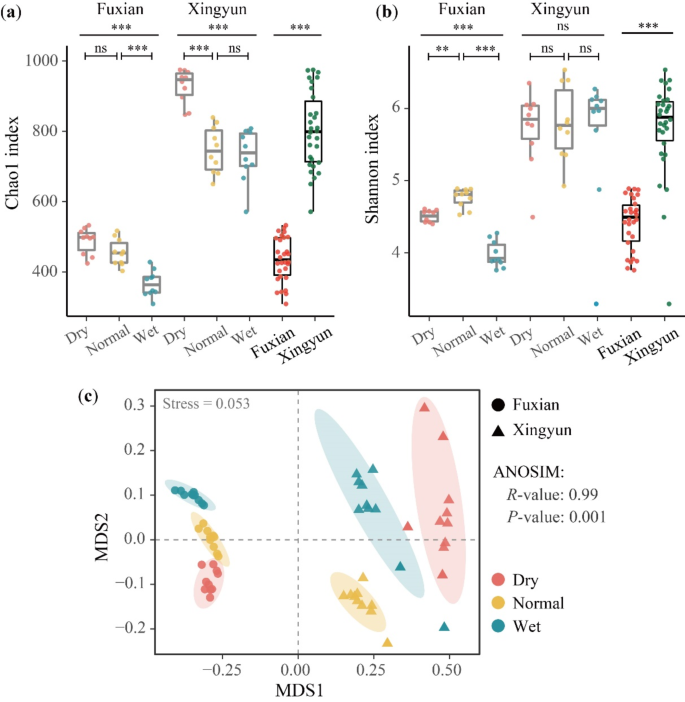
Comparisons of bacterial α-diversity and β-diversity between the oligotrophic Lake Fuxia and hypereutrophic Lake Xingyun, as well as among different hydrological periods. ( a ) Chao1 index, ( b ) Shannon index. ***, P < 0.001; **, P < 0.01; ns, no significant. ( c ) Nonmetric multidimensional scaling (NMDS) plot based on Bray–Curtis dissimilarity. Differences between bacterial community structures between the two lakes were tested using Analysis of Similarities (ANOSIM). The results are presented in the NMDS plot. Ellipses cover 95% of the data for each hydrological period
Taxonomy of bacterial communities
At the phylum level, Lake Fuxian exhibited the presence of 22 bacterial phyla, while Lake Xingyun showed 23. Notably, Actinobacteria stood out as the most abundant phylum in Lake Fuxian, with an average relative abundance exceeding 79.0%. In contrast, Lake Xingyun displayed Actinobacteria (32.0%), Cyanobacteria (23.9%), Proteobacteria (21.0%), and Bacteroidetes (20.0%) as the most abundant phyla (Fig. 4 a). A significant shift in bacterial taxa occurred during the wet season in Lake Fuxian, characterized by a substantial decrease in the relative abundance of Proteobacteria and a remarkable increase in Cyanobacteria.
At the class level, Lake Fuxian revealed the presence of 43 bacterial classes, whereas Lake Xingyun exhibited 47. Acidimicrobiia (54.0%) and Actinobacteria (25.2%) dominated Lake Fuxian, while Lake Xingyun was characterized by Actinobacteria (25.2%), Oxyphotobacteria (23.8%), Bacteroidia (17.2%), Gammaproteobacteria (14.9%), and Alphaproteobacteria (10.8%) (Fig. 4 b).
Moving to the genus level, Lake Fuxian was dominated by the CL500-29 marine group (52.7%), hgcI clade (19.8%), and Cyanobium (4.9%). Conversely, Lake Xingyun featured Microcystis (19.9%), hgcI clade (15.7%), CL500-29 marine group (6.6%), Flavobacterium (4.3%), and unclassified Microscillaceae (4.3%) (Supplementary Fig. S1 & Table S2 ).
To discern differences in bacterial taxonomy between Lake Fuxian and Lake Xingyun, LEfSe was employed. At the phylum level, LEfSe revealed a significant enrichment of Actinobacteria in Lake Fuxian, whereas Bacteroidetes, Cyanobacteria, and Proteobacteria were notably enriched in Lake Xingyun (Fig. 4 c). At the family level, substantial differences were observed, including Ilumatobacteria and Cyanobiaceae in Lake Fuxian, and Microscillaceae, Flavobacteriaceae, Microcystaceae, Acetobacteraceae, and Burkholderiaceae in Lake Xingyun (Fig. 4 c).
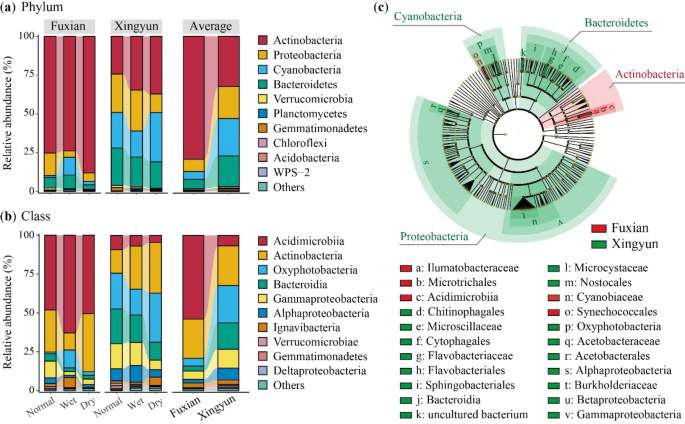
Bacterial taxonomy of Lake Fuxian and Lake Xingyun, categorized at the ( a ) phylum and ( b ) class levels. Only the 10 most abundant taxa are included in the figure, while other rare taxa are grouped into “Others”. ( c ) The LEfSe cladogram shows significant differences in bacterial taxa between the two lakes. Colored dots on the cladogram denote taxa with noteworthy differences in abundance across lakes, while the cladogram circles delineate phylogenetic taxa from phylum to family
Environmental drivers influencing BCCs
RDA unveiled that the seasonal variations in BCCs between Lakes Fuxian and Xingyun were primarily influenced by COD Mn , DOC, PO 4 -P, NH 4 -N, WT, and DO (Fig. 5 a). Together, these six variables accounted for 80.2% of the total variations, with COD Mn and DOC emerging as the most crucial contributors (Fig. 5 b). In Lake Fuxian, the first and second axes of the RDA explained 24.3% and 15.4% of the variance in bacterial community (Fig. 5 c). Through forward selection of RDA and the Monte Carlo permutation test, it was determined that the variations in BCCs in Lake Fuxian were significantly driven by DOC, PO 4 -P, TDP, TN, and NO 3 -N. Similarly, in Lake Xingyun, the first and second axes of the RDA explained 27.7% and 21.3% of the variance in BCCs (Fig. 5 d). The forward selection of RDA, along with the Monte Carlo permutation test, indicated that the variations in BCCs in Lake Xingyun could be significantly elucidated by Cond, NO 3 -N, pH, PO 4 -P, and WT.
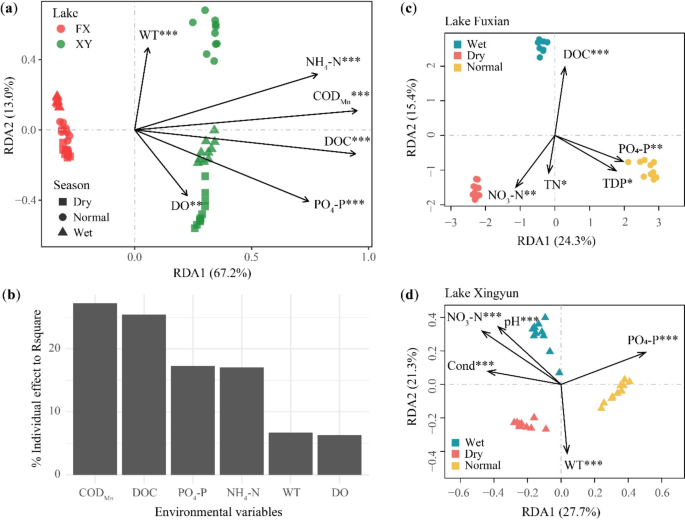
Redundancy analyses (RDA) plots depict the prominent environmental varibles influencing variations in BCCs in both lakes ( a ) and within each specific lake, namely Lake Fuxian ( c ) and Lake Xingyun ( d ). The individual effects of each significant environmental variable are illustrated in figure ( b ). The significance levels: * P < 0.05, ** P < 0.01, *** P < 0.001. The abbreviations used in the plots are as follows: COD Mn , permanganate index; DOC, dissolved organic carbon; PO 4 -P, phosphate phosphorus; NH 4 -N, ammonia nitrogen; WT, water temperature; DO, dissolved oxygen; NO 3 -N, nitrate nitrogen; TN, total nitrogen; TDP, total dissolved phosphorus; Cond, conductivity
Functional prediction analysis
The results of functional prediction revealed notable differences in the abundance of various potential functions between Lake Fuxian and Lake Xingyun (Fig. 6 ). In Lake Fuxian, functions such as intracellular parasites, methanol oxidation, methylotrophy, fermentation, photoheterotrophy, and aromatic compound degradation were found to be more abundant. Additionally, functions associated with the nitrogen cycle, including nitrogen/nitrate respiration and nitrogen/nitrate denitrification, were enriched in Lake Fuxian. Conversely, Lake Xingyun exhibited significant enrichments in functions related to ureolysis, human pathogens, animal parasites or symbionts, and phototrophy.
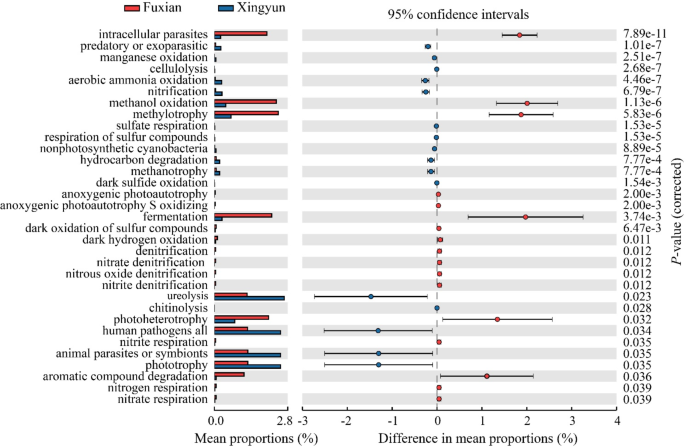
Mean proportion of bacterial functional groups with the significant difference ( P < 0.05) between Lake Fuxian and Lake Xingyun
Characteristics of co-occurrence networks
Co-occurrence networks were constructed based on datasets from all hydrological seasons, and the observed empirical network parameters significantly surpassed those of the corresponding random networks, indicating a nonrandom nature (Table 1 ). Moreover, the “small-world” coefficients (σ) indicated that both co-occurrence networks exhibited “small-world” properties, suggesting their ability to respond rapidly and effectively to disturbances [ 31 ]. Additionally, both lakes’ co-occurrence networks displayed predominantly positive relationships, with higher values in Lake Fuxian (71.6%) compared to Lake Xingyun (67.4%), indicative of ecological cooperation within the microbiome.
Contrasting the oligotrophic Lake Fuxian, co-occurrence networks from the hypereutrophic Lake Xingyun exhibited higher avgK (9.330 vs. 4.639) and avgCC (0.415 vs. 0.371), indicating increased complexity. The Lake Xingyun network demonstrated higher robustness and lower vulnerability, suggesting a more stable characteristic. Conversely, the co-occurrence network from Lake Fuxian displayed a shorter GD, implying greater sensitivity to environmental disturbances. Both lakes’ co-occurrence networks exhibited modular structures, clearly divided into four major modules (Fig. 7 ). In Lake Fuxian, modules I were predominantly present in the wet season, while modules IV were mainly observed in the dry season. In Lake Xingyun, modules I and III were primarily associated with the wet season.
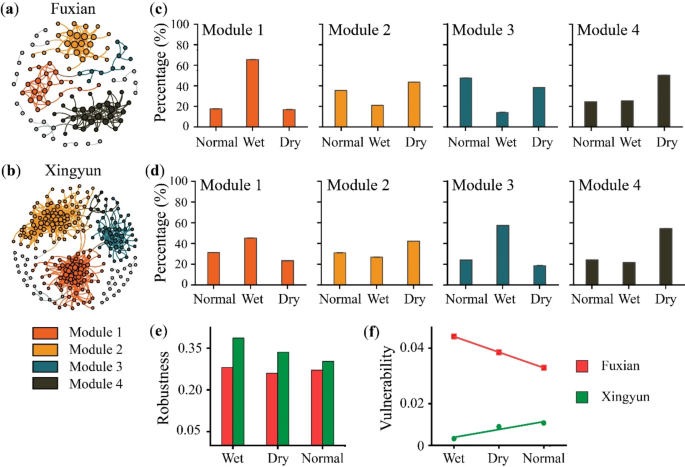
Co-occurrence networks and their topological properties and stability of bacterial communities from Lake Fuxian ( a ) and Lake Xingyun ( b ). ASVs were selected based on their relative abundance (≥ 0.08%) among the total bacterial sequences. The size of each node is proportional to the number of connections to that node, and different colors indicate distinct modules within the networks. The distribution of bacterial taxa in each module from Lake Fuxian ( c ) and Lake Xingyun ( d ) is presented across different hydrological periods. To assess network stability, robustness ( e ) was quantified as the proportion of remaining taxa in a network after random removal of node (50%). Vulnerability ( f ) was determined by identifying the maximum node vulnerability in each network
Building upon our primary objectives outlined in the introduction, we explored the distinct differences in bacterial diversity, taxonomic composition, and predicted functions between the oligotrophic Lake Fuxian and the hypereutrophic Lake Xingyun. As hypothesized, our findings revealed significant disparities in the bacterial communities of these lakes, driven by their contrasting nutrient states. The results provide valuable insights into how environmental factors, specifically trophic conditions, shape bacterial community composition and ecological interactions within freshwater ecosystems. The following, we will discuss in detail from three aspects.
Distinct differences in bacterial diversity, taxonomic composition and predicted functions between oligotrophic Lake Fuxian and hypereutrophic Lake Xingyun
Our exploration of bacterial diversity, taxonomic composition, and functional predictions in oligotrophic Lake Fuxian and hypereutrophic Lake Xingyun reveals distinct disparities (Fig. 3 a, b). The bacterial communities in the hypereutrophic lake exhibit considerably greater α-diversity than those in the oligotrophic counterpart. Our results suggest that in hyper-eutrophic lakes, the increased availability of growth-limiting resources may lead to the diversification of bacterial niches, which could enhance species diversity at the local scale [ 49 , 50 ]. Additionally, due to its small surface area and shallow depth, Lake Xingyun is more susceptible to external river inputs. Therefore, the influx of external microorganisms may also be one of the reasons for the high bacterial alpha diversity in Lake Xingyun.
Despite both lakes sharing common bacterial phyla with other freshwater bodies [ 10 , 51 ], their relative abundances diverge significantly, illustrating how eutrophication alters bacterial diversity and community structures [ 52 ]. In Lake Fuxian, Actinobacteria dominate the bacterial community, comprising 79.0% of its composition. This phylum, widespread in various aquatic environments, has traditionally been associated with different water bodies [ 7 ], including mesotrophic drinking water reservoirs [ 53 , 54 ], oligotrophic and eutrophic lakes [ 55 , 56 , 57 ]. Mesocosm experiments revealed that the occurrence of Actinobacteria was correlated with less eutrophic conditions [ 58 ]. Our LEfSe analysis further emphasizes the significant enrichment of Actinobacteria in Lake Fuxian compared to Lake Xingyun, suggesting an adaptation to the lake’s upper water layers from oligotrophic environments [ 58 ]. Conversely, Lake Xingyun exhibits higher enrichment of Proteobacteria, Cyanobacteria, and Bacteroidetes, attributed to its heightened nutrient sensitivity and human-induced waste contributions. This hypereutrophic lake, characterized by significant human disturbance, heavily relies on precipitation and inflow from surrounding rivers, intensifying its exposure to agricultural runoff [ 34 ]. Proteobacteria and Bacteroidetes exhibit high sensitivity to nutrient enrichment and cyanobacterial blooms, evident in their prevalence in environments with such conditions [ 59 , 60 ]. Additionally, Bacteroidetes have been linked to areas with elevated nitrogen levels and the presence of human metabolic wastes in lakes [ 61 ], highlighting their adaptability to nutrient-rich and anthropogenically influenced aquatic ecosystems.
Significant differences in functional composition between the two trophic lakes were observed. In Lake Fuxian, more functions related to element cycling were identified, while Lake Xingyun exhibited a higher abundance of functions related to human interference. In Lake Fuxian, functions associated with carbon decomposition (fermentation, and aromatic compound degradation) were significantly enriched, with the dominant bacterial genus CL500-29 marine group and hgcI clade showing high efficiency in utilizing carbon compounds [ 62 , 63 , 64 ]. Furthermore, functions related to methanol utilization (methanol oxidation, and methylotrophy) in Lake Fuxian were also significantly enriched. This is consistent with the fact that the relative abundance of methylotrophs (i.e., Candidatus Methylopumilus , also known as the freshwater LD28 tribe) [ 65 , 66 ] in Lake Fuxian is more than twice that in Lake Xingyun (Supplementary Table S2 ). Effects related to methyl nutrition play a role in the global nitrogen cycle [ 65 ], with methanol enhancing denitrification. Functions related to the nitrogen cycle (nitrogen/nitrate respiration, nitrogen/nitrate denitrification) were enriched in Lake Fuxian. Despite the hypereutrophic lake being rich in nitrogen, extreme hypoxia under high algal biomass significantly restricted nitrification, consequently limiting denitrification due to the lack of available substrates [ 67 ]. Additionally, the enrichment of functions related to ureolysis, human pathogens, and animal parasites or symbionts in Lake Xingyun suggests the presence of exogenous nitrogen input from agricultural non-point sources and other pollutants, posing a potential pathogenic risk for surrounding residents.
Nutrients are the primary factor driving differences in BCCs between Lake Fuxian and Lake Xingyun
Environmental factors exert significant influence on BCCs in Lakes Fuxian and Xingyun. The intricate interplay between the water environment and microorganisms plays a vital role in shaping microbial distribution and ecological functions [ 68 , 69 ]. Results from the RDA analysis highlight the predominant impact of nutrient-related factors (e.g., COD Mn , DOC, PO 4 -P and NH 4 -N) compared to physical factors (e.g., WT and DO) on the variations in BCCs between the two lakes (Fig. 5 ). Carbon, nitrogen, and phosphorus emerge as crucial determinants affecting bacterial community composition, providing the material foundation for bacterial growth and reproduction in aquatic environments [ 70 ].
The commonly used COD Mn serves as an indicator of organic pollution levels in water, with its concentration significantly influencing microbial community structures in lakes [ 71 , 72 ]. DOC, which influences the microbial carbon cycle through its effects on microbial respiration and metabolism, plays a pivotal role in shaping bacterial communities [ 73 , 74 ]. Studies on DOC sources in lakes across different trophic levels suggest that oligotrophic lakes primarily receive endogenous DOC inputs, while eutrophic lakes benefit from both exogenous and endogenous carbon sources [ 75 ]. The profound impact of DOC on bacterial community composition is particularly evident in Lake Fuxian (Fig. 5 c), where bacteria with high carbon compound utilization, such as the CL500-29 marine group and hgcI clade , dominate, comprising up to 80.3% of the community during the dry season (Supplementary Fig. S1 ). The oligotrophic conditions of Lake Fuxian emphasize the importance of carbon in shaping bacterial community composition, given the limited availability of organic carbon sources.
Excessive nutrient levels, particularly nitrogen and phosphorus, promote the growth of algae, including cyanobacteria. In eutrophic Lake Xingyun, the growth of abundant Cyanobacteria, mainly Microcystis , is influenced by dissolved nitrogen and phosphorus elements. Cyanobacteria may compete with heterotrophic bacteria for these nutrients during their growth, establishing a cooperative relationship by recruiting beneficial heterotrophic bacteria through the release of extracellular polysaccharides or DOC [ 76 , 77 ]. Additionally, the proliferation of cyanobacteria significantly increases water pH, affecting nutrient solubility and bioavailability [ 78 ], thereby influencing bacterial community dynamics. Bacterial degradation of organic matter further promotes the regeneration of nitrogen and phosphorus nutrients, stimulating cyanobacterial growth [ 79 ]. In summary, the complex interplay between algae and bacteria plays a pivotal role in maintaining the dynamic balance of bacterial communities [ 76 , 80 ].
While our study primarily focuses on the impact of nutrient conditions on bacterial community structure in hypereutrophic Lake Xingyun and oligotrophic Lake Fuxian, it does not extensively address the role of protists and viruses. These microorganisms influence bacterial communities through predation and viral lysis (top-down control) and nutrient regeneration (bottom-up control) [ 81 , 82 ]. For example, the dominance of the hgcI clade in both lakes in this study (Supplementary Fig. S1 ) may be related to their adaptive life strategies, which include responses to top-down mechanisms and efficient resource utilization [ 83 ]. The varying impacts of these processes in different trophic environments are significant and warrant further investigation. Future studies should incorporate these top-down forces to provide a more comprehensive understanding of bacterial community dynamics in freshwater lakes.
Bacterial networks in hypereutrophic Lake Xingyun are more complex and stable than in oligotrophic Lake Fuxian
Our findings reveal a higher level of complexity in bacterial networks within the hypereutrophic Lake Xingyun compared to the oligotrophic Lake Fuxian (Table 1 ; Fig. 7 ). This aligns with a recent study covering oligotrophic to hypereutrophic lakes, which demonstrated a unimodal pattern in bacterioplankton network complexity across increasing trophic state index, peaking at mesotrophic states and subsequently decreasing towards hyper-eutrophic states [ 17 ]. Our results support this trend by confirming that bacterial networks are more intricate in hypereutrophic lakes compared to their oligotrophic counterparts.
In recent decades, numerous studies have explored the stability of ecosystems by examining interaction network structures [ 23 , 84 ]. Prior research has consistently shown a robust positive correlation between network complexity and stability [ 31 ], aligning with the macroecological perspective that increased ecosystem complexity leads to enhanced stability [ 23 ]. Conversely, in random networks, an elevation in modularity correlates with a higher likelihood of network instability, establishing a negative correlation between the two [ 85 , 86 ]. Our results indicate a higher modularity in bacterial networks in oligotrophic states (modularity: 0.702 vs. 0.612), predicting a more stable network in hypereutrophic states. Correspondingly, calculations for stability (robustness and vulnerability) support this outcome (Fig. 7 ). The shorter average path length of the bacterial network in Lake Fuxian suggests greater efficiency in information, energy, and material transfer between species [ 31 ]. This implies that the bacterial community in Lake Fuxian can respond more rapidly to perturbations, rendering the community more prone to changes [ 87 ]. Therefore, the higher network complexity in hypereutrophic lakes suggests that bacterial stability in such environments may surpass that in oligotrophic lakes. Consequently, oligotrophic lakes may possess weaker resistance to environmental disturbances, rendering their ecosystems more vulnerable.
The response of bacterial species to various perturbations determines community stability, with the average degree reflecting the complex relationships between species interactions [ 88 ]. The higher average degree in the hypereutrophic lake indicates more intricate interactions between bacterial communities in Lake Xingyun. Additionally, the heightened negative correlations in bacterial networks in the hypereutrophic state suggest that bacterial communities in Lake Xingyun engage in more competitive relationships with each other. As a typical algal-type lake, the bacteria-algae system in Lake Xingyun forms complex interrelationships, including mutualistic cooperation and competitive exclusion [ 25 ], making it more resistant to environmental disturbances. Despite being a hypereutrophic lake experiencing year-round cyanobacterial bloom accumulation [ 89 ], coupled with the stability of bacterial communities, the ecological restoration of Lake Xingyun’s ecosystem from a turbid water algal state to a clear water macrophyte state becomes a more challenging endeavor.
Conclusions
In this study, we examined the bacterial communities in two plateau freshwater lakes with contrasting trophic states—oligotrophic and hypereutrophic—in Yunnan, China. Our results revealed that the hypereutrophic lake exhibited higher α-diversity and β-diversity compared to the oligotrophic lake. Distinct differences in bacterial community compositions were observed between the two ecosystems. In the oligotrophic lake, bacterial communities were primarily involved in carbon and nitrogen cycling processes, while in the hypereutrophic lake, greater human activity-related disturbances highlighted potential pathogenic risks. Additionally, the bacterial community network in the hypereutrophic lake was more complex and stable, suggesting challenges for ecological restoration in lakes with severe cyanobacterial blooms.
Data availability
The raw sequence data reported in this paper have been archived in the Genome Sequence Archive at the BIG Data Center (http://bigd.big.ac.cn/gsa) under the accession number CRA008654.
Adrian R, O’Reilly CM, Zagarese H, Baines SB, Hessen DO, Keller W, et al. Lakes as sentinels of climate change. Limnol Oceanogr. 2009;54(6):2283–97. https://doi.org/10.4319/lo.2009.54.6_part_2.2283 .
Article PubMed PubMed Central Google Scholar
Adrian R, Hessen DO, Blenckner T, Hillebrand H, Hilt S, Jeppesen E, et al. Environmental impacts—Lake ecosystems. In: Quante M, Colijn F, editors. North Sea Region Climate Change Assessment. Cham: Springer International Publishing; 2016. pp. 315–40.
Chapter Google Scholar
Schindler DW. Lakes as sentinels and integrators for the effects of climate change on watersheds, airsheds, and landscapes. Limnol Oceanogr. 2009;54(6):2349–58.
Article CAS Google Scholar
Zhang X, Li B, Xu H, Wells M, Tefsen B, Qin B. Effect of micronutrients on algae in different regions of Taihu, a large, spatially diverse, hypereutrophic lake. Water Res. 2019;151:500–14. https://doi.org/10.1016/j.watres.2018.12.023 .
Article CAS PubMed Google Scholar
Ding J, Cao J, Xu Q, Xi B, Su J, Gao R, et al. Spatial heterogeneity of lake eutrophication caused by physiogeographic conditions: an analysis of 143 lakes in China. J Environ Sci. 2015;30:140–7. https://doi.org/10.1016/j.jes.2014.07.029 .
Wang Y, Guo M, Li X, Liu G, Hua Y, Zhao J, et al. Shifts in microbial communities in shallow lakes depending on trophic states: feasibility as an evaluation index for eutrophication. Ecol Indic. 2022;136:108691. https://doi.org/10.1016/j.ecolind.2022.108691 .
Article Google Scholar
Newton RJ, Jones SE, Eiler A, McMahon KD, Bertilsson S. A guide to the natural history of freshwater lake bacteria. Microbiol Mol Biol Rev. 2011;75(1):14–49. doi: Doi 10.1128/Mmbr.00028 – 10.
Article CAS PubMed PubMed Central Google Scholar
Tandon K, Yang S-H, Wan M-T, Yang C-C, Baatar B, Chiu C-Y, et al. Bacterial community in water and air of two sub-alpine lakes in Taiwan. Microbes Environ. 2018;33(2):120–6. https://doi.org/10.1264/jsme2.ME17148 .
Shang Y, Wu X, Wang X, Wei Q, Ma S, Sun G, et al. Factors affecting seasonal variation of microbial community structure in Hulun Lake, China. Sci Total Environ. 2022;805:150294. https://doi.org/10.1016/j.scitotenv.2021.150294 .
Ji B, Liang J, Ma Y, Zhu L, Liu Y. Bacterial community and eutrophic index analysis of the East Lake. Environ Pollut. 2019;252:682–8. https://doi.org/10.1016/j.envpol.2019.05.138 .
Chrost RJ, Koton M, Siuda W. Bacterial secondary production and bacterial biomass in four mazurian lakes of differing trophic status. Pol J Environ Stud. 2000;9(4):255–66.
CAS Google Scholar
Feng C, Jia J, Wang C, Han M, Dong C, Huo B, et al. Phytoplankton and bacterial community structure in two Chinese lakes of different trophic status. Microorganisms. 2019;7(12):621. https://doi.org/10.3390/microorganisms7120621 .
Huang W, Chen X, Jiang X, Zheng BH. Characterization of sediment bacterial communities in plain lakes with different trophic statuses. Microbiologyopen. 2017;6(5). https://doi.org/10.1002/mbo3.503 .
Ren Z, Qu XD, Peng WQ, Yu Y, Zhang M. Functional properties of bacterial communities in water and sediment of the eutrophic river-lake system of Poyang Lake, China. Peerj. 2019;7. https://doi.org/10.7717/peerj.7318 .
Kiersztyn B, Chróst R, Kaliński T, Siuda W, Bukowska A, Kowalczyk G, et al. Structural and functional microbial diversity along a eutrophication gradient of interconnected lakes undergoing anthropopressure. Sci Rep. 2019;9(1):11144. https://doi.org/10.1038/s41598-019-47577-8 .
Yang W, Zheng C, Zheng Z, Wei Y, Lu K, Zhu J. Nutrient enrichment during shrimp cultivation alters bacterioplankton assemblies and destroys community stability. Ecotox Environ Safe. 2018;156:366–74. https://doi.org/10.1016/j.ecoenv.2018.03.043 .
Shen Z, Xie G, Yu B, Zhang Y, Shao K, Gong Y, et al. Eutrophication diminishes bacterioplankton functional dissimilarity and network complexity while enhancing stability: implications for the management of eutrophic lakes. J Environ Manage. 2024;352:120119. https://doi.org/10.1016/j.jenvman.2024.120119 .
Paerl HW, Xu H, McCarthy MJ, Zhu G, Qin B, Li Y, et al. Controlling harmful cyanobacterial blooms in a hyper-eutrophic lake (Lake Taihu, China): the need for a dual nutrient (N & P) management strategy. Water Res. 2011;45(5):1973–83. https://doi.org/10.1016/j.watres.2010.09.018 .
Ta Dang T, Bui Quoc L, Le Minh T, Harada M, Hibamatsu K, Tabata T. Eutrophication status of lakes in Inner Hanoi and a case study of Cu Chinh Lake. J Fac Agric Kyushu Univ. 2021;66(1):97–104.
Google Scholar
Xie G, Tang X, Gong Y, Shao K, Gao G. How do planktonic particle collection methods affect bacterial diversity estimates and community composition in oligo-, meso- and eutrophic lakes? Front Microbiol. 2020;11:593589. https://doi.org/10.3389/fmicb.2020.593589 .
Yanez-Montalvo A, Aguila B, Gómez-Acata ES, Guerrero-Jacinto M, Oseguera LA, Falcón LI, et al. Shifts in water column microbial composition associated to lakes with different trophic conditions: Lagunas De Montebello National Park, Chiapas, México. PeerJ. 2022;10:e13999. https://doi.org/10.7717/peerj.13999 .
Ji B, Qin H, Guo S, Chen W, Zhang X, Liang J. Bacterial communities of four adjacent fresh lakes at different trophic status. Ecotoxicol Environ Saf. 2018;157:388–94. https://doi.org/10.1016/j.ecoenv.2018.03.086 .
Montoya JM, Pimm SL, Sole RV. Ecological networks and their fragility. Nature. 2006;442(7100):259–64. https://doi.org/10.1038/nature04927 .
Mougi A, Kondoh M. Diversity of interaction types and ecological community stability. Science. 2012;337(6092):349–51. https://doi.org/10.1126/science.1220529 .
Faust K, Raes J. Microbial interactions: from networks to models. Nat Rev Microbiol. 2012;10(8):538–50. https://doi.org/10.1038/nrmicro2832 .
Allesina S, Levine JM. A competitive network theory of species diversity. Proc Natl Acad Sci U S A. 2011;108(14):5638–42. https://doi.org/10.1073/pnas.1014428108 .
Huelsmann M, Ackermann M. Community instability in the microbial world. Science. 2022;378(6615):29–30. https://doi.org/10.1126/science.ade2516 .
Jiao S, Lu YH, Wei GH. Soil multitrophic network complexity enhances the link between biodiversity and multifunctionality in agricultural systems. Glob Change Biol. 2022;28(1):140–53. https://doi.org/10.1111/gcb.15917 .
Kara EL, Hanson PC, Hu YH, Winslow L, McMahon KD. A decade of seasonal dynamics and co-occurrences within freshwater bacterioplankton communities from eutrophic Lake Mendota, WI, USA. ISME J. 2013;7(3):680–4. https://doi.org/10.1038/ismej.2012.118 .
Article PubMed Google Scholar
Mo Y, Peng F, Jeppesen E, Gamfeldt L, Xiao P, Al MA, et al. Microbial network complexity drives non-linear shift in biodiversity-nutrient cycling in a saline urban reservoir. Sci Total Environ. 2022;850:158011. https://doi.org/10.1016/j.scitotenv.2022.158011 .
Yuan MM, Guo X, Wu L, Zhang Y, Xiao N, Ning D, et al. Climate warming enhances microbial network complexity and stability. Nat Clim Chang. 2021;11(4):343–8. https://doi.org/10.1038/s41558-021-00989-9 .
Xing P, Tao Y, Luo J, Wang L, Li B, Li H, et al. Stratification of microbiomes during the holomictic period of Lake Fuxian, an alpine monomictic lake. Limnol Oceanogr. 2020;65(S1):S134–48. https://doi.org/10.1002/lno.11346 .
Shen M, Li Q, Ren M, Lin Y, Wang J, Chen L, et al. Trophic status is associated with community structure and metabolic potential of planktonic microbiota in plateau lakes. Front Microbiol. 2019;10. https://doi.org/10.3389/fmicb.2019.02560 .
Chen X, Huang X, Wu D, Chen J, Zhang J, Zhou A, et al. Late Holocene land use evolution and vegetation response to climate change in the watershed of Xingyun Lake, SW China. CATENA. 2022;211:105973. https://doi.org/10.1016/j.catena.2021.105973 .
Jin X, Tu Q. Specification for Lake Eutrophication Investigation: 2nd Edition. Beijing: China Environmental Science Press; 1990.
Gong Y, Tang X, Shao K, Hu Y, Gao G. Dynamics of bacterial abundance and the related environmental factors in large shallow Eutrophic Lake Taihu. J Freshw Ecol. 2017;32(1):133–45. https://doi.org/10.1080/02705060.2016.1248506 .
Lin S-S, Shen S-L, Zhou A, Lyu H-M. Assessment andmanagement of lake eutrophication: a case study in Lake Erhai, China. Sci Total Environ. 2021;751. https://doi.org/10.1016/j.scitotenv.2020.141618 .
Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM, et al. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome. 2014;2. https://doi.org/10.1186/2049-2618-2-6 .
Bolyen E, Rideout JR, Dillon MR, Bokulich N, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37(8):852–7. https://doi.org/10.1038/s41587-019-0209-9 .
Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome. 2018;6(1):90. https://doi.org/10.1186/s40168-018-0470-z .
Clarke KR. Non-parametric multivariate analyses of changes in community structure. Aust J Ecol. 1993;18(1):117–43. https://doi.org/10.1111/j.1442-9993.1993.tb00438.x .
Lai J, Zou Y, Zhang J, Peres-Neto PR. Generalizing hierarchical and variation partitioning in multiple regression and canonical analyses using the rdacca.hp R package. Methods Ecol Evol. 2022;13(4):782–8. https://doi.org/10.1111/2041-210X.13800 .
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. https://doi.org/10.1186/gb-2011-12-6-r60 .
Louca S, Parfrey LW, Doebeli M. Decoupling function and taxonomy in the global ocean microbiome. Science. 2016;353(6305):1272–7. https://doi.org/10.1126/science.aaf4507 .
Feng K, Peng X, Zhang Z, Gu S, He Q, Shen W et al. iNAP: An integrated network analysis pipeline for microbiome studies. 2022;1(2):e13; https://doi.org/10.1002/imt2.13
Newman MEJ. Fast algorithm for detecting community structure in networks. Phys Rev E. 2004;69(6). https://doi.org/10.1103/PhysRevE.69.066133 .
Bastian M, Heymann S, Jacomy M, Gephi. An Open Source Software for Exploring and Manipulating Networks. 2009.
Lopes CT, Franz M, Kazi F, Donaldson SL, Morris Q, Bader GD. Cytoscape web: an interactive web-based network browser. Bioinformatics. 2010;26(18):2347–8. https://doi.org/10.1093/bioinformatics/btq430 .
Eiler A, Bertilsson S. Flavobacteria blooms in four eutrophic lakes: linking population dynamics of freshwater bacterioplankton to resource availability. Appl Environ Microbiol. 2007;73(11):3511–8. https://doi.org/10.1128/Aem.02534-06 .
Bai L, Cao C, Wang C, Xu H, Zhang H, Slaveykova VI, et al. Toward quantitative understanding of the bioavailability of dissolved organic matter in freshwater lake during cyanobacteria blooming. Environ Sci Technol. 2017;51(11):6018–26. https://doi.org/10.1021/acs.est.7b00826 .
Rummens K, De Meester L, Souffreau C. Inoculation history affects community composition in experimental freshwater bacterioplankton communities. Environ Microbiol. 2018;20(3):1120–33. https://doi.org/10.1111/1462-2920.14053 .
Hiller KA, Foreman KH, Weisman D, Bowen JL. Permeable reactive barriers designed to mitigate eutrophication alter bacterial community composition and aquifer redox conditions. Appl Environ Microbiol. 2015;81(20):7114–24. https://doi.org/10.1128/AEM.01986-15 .
Zhang H, Ma M, Huang T, Miao Y, Li H, Liu K, et al. Spatial and temporal dynamics of actinobacteria in drinking water reservoirs: novel insights into abundance, community structure, and co-existence model. Sci Total Environ. 2022;814:152804. https://doi.org/10.1016/j.scitotenv.2021.152804 .
Yu B, Xie G, Shen Z, Shao K, Tang X. Spatiotemporal variations, assembly processes, and co-occurrence patterns of particle-attached and free-living bacteria in a large drinking water reservoir in China. Front Microbiol. 2023;13:1056147. https://doi.org/10.3389/fmicb.2022.1056147 .
Xie G, Martin RM, Liu C, Zhang L, Tang X. Patterns of free-living and particle-attached bacteria along environmental gradients in Lake Taihu. Can J Microbiol. 2023;69(6):228–39. https://doi.org/10.1139/cjm-2022-0243 .
Shen Z, Xie G, Zhang Y, Yu B, Shao K, Gao G, et al. Similar assembly mechanisms but distinct co-occurrence patterns of free-living vs. particle-attached bacterial communities across different habitats and seasons in shallow, eutrophic Lake Taihu. Environ Pollut. 2022;314:120305. https://doi.org/10.1016/j.envpol.2022.120305 .
Chao J, Li J, Kong M, Shao K, Tang X. Bacterioplankton diversity and potential health risks in volcanic lakes: a study from Arxan Geopark, China. Environ Pollut. 2024;342:123058. https://doi.org/10.1016/j.envpol.2023.123058 .
Haukka K, Kolmonen E, Hyder R, Hietala J, Vakkilainen K, Kairesalo T, et al. Effect of nutrient loading on bacterioplankton community composition in lake mesocosms. Microb Ecol. 2006;51(2):137–46. https://doi.org/10.1007/s00248-005-0049-7 .
Bernhard AE, Colbert D, McManus J, Field KG. Microbial community dynamics based on 16S rRNA gene profiles in a Pacific Northwest estuary and its tributaries. FEMS Microbiol Ecol. 2005;52(1):115–28. https://doi.org/10.1016/j.femsec.2004.10.016 .
Scherer PI, Millard AD, Miller A, Schoen R, Raeder U, Geist J, et al. Temporal dynamics of the microbial community composition with a focus on toxic cyanobacteria and toxin presence during harmful algal blooms in two south German lakes. Front Microbiol. 2017;8:2387. https://doi.org/10.3389/fmicb.2017.02387 .
Zhang L, Zhong M, Li X, Lu W, Li J. River bacterial community structure and co-occurrence patterns under the influence of different domestic sewage types. J Environ Manage. 2020;266:110590. https://doi.org/10.1016/j.jenvman.2020.110590 .
Shi P, Wang H, Feng M, Cheng H, Yang Q, Yan Y, et al. The coupling response between different bacterial metabolic gunctions in water and sediment improve the ability to mitigate climate change. Water. 2022;14(8). https://doi.org/10.3390/w14081203 .
Lindh MV, Lefebure R, Degerman R, Lundin D, Andersson A, Pinhassi J. Consequences of increased terrestrial dissolved organic matter and temperature on bacterioplankton community composition during a Baltic Sea mesocosm experiment. Ambio. 2015;44:S402–12. https://doi.org/10.1007/s13280-015-0659-3 .
Ghylin TW, Garcia SL, Moya F, Oyserman BO, Schwientek P, Forest KT, et al. Comparative single-cell genomics reveals potential ecological niches for the freshwater acl Actinobacteria lineage. ISME J. 2014;8(12):2503–16. https://doi.org/10.1038/ismej.2014.135 .
Kalyuhznaya MG, Martens-Habbena W, Wang T, Hackett M, Stolyar SM, Stahl DA, et al. Methylophilaceae link methanol oxidation to denitrification in freshwater lake sediment as suggested by stable isotope probing and pure culture analysis. Environ Microbiol Rep. 2009;1(5):385–92. https://doi.org/10.1111/j.1758-2229.2009.00046.x .
Ramachandran A, Walsh DA. Investigation of XoxF methanol dehydrogenases reveals new methylotrophic bacteria in pelagic marine and freshwater ecosystems. FEMS Microbiol Ecol. 2015;91(10). https://doi.org/10.1093/femsec/fiv105 .
Yao Y, Liu H, Han R, Li D, Zhang L. Identifying the mechanisms behind the positive feedback loop between nitrogen cycling and algal blooms in a shallow eutrophic lake. Water. 2021;13(4):524. https://doi.org/10.3390/w13040524 .
Feng J, Zhou L, Zhao X, Chen J, Li Z, Liu Y, et al. Evaluation of environmental factors and microbial community structure in an important drinking-water reservoir across seasons. Front Microbiol. 2023;14:1091818.
Tang X, Xie G, Shao K, Tian W, Gao G, Qin B. Aquatic bacterial diversity, community composition and assembly in the semi-arid Inner Mongolia Plateau: combined effects of salinity and nutrient levels. Microorganisms. 2021;9(2):208. https://doi.org/10.3390/microorganisms9020208 .
Zwirglmaier K, Keiz K, Engel M, Geist J, Raeder U. Seasonal and spatial patterns of microbial diversity along a trophic gradient in the interconnected lakes of the Osterseen Lake District, Bavaria. Front Microbiol. 2015;6. https://doi.org/10.3389/fmicb.2015.01168 .
Zhou S, Sun Y, Yu M, Shi Z, Zhang H, Peng R, et al. Linking shifts in bacterial community composition and function with changes in the dissolved organic matter pool in ice-covered Baiyangdian Lake, Northern China. Microorganisms. 2020;8(6):883. https://doi.org/10.3390/microorganisms8060883 .
Guo J, Zheng Y, Teng J, Song J, Wang X, Zhao Q. The seasonal variation of microbial communities in drinking water sources in Shanghai. J Clean Prod. 2020;265:121604. https://doi.org/10.1016/j.jclepro.2020.121604 .
Dong H, Zhang S, Lin J, Zhu B. Responses of soil microbial biomass carbon and dissolved organic carbon to drying-rewetting cycles: a meta-analysis. CATENA. 2021;207:105610. https://doi.org/10.1016/j.catena.2021.105610 .
Fonseca BM, Levi EE, Jensen LW, Graeber D, Søndergaard M, Lauridsen TL, et al. Effects of DOC addition from different sources on phytoplankton community in a temperate eutrophic lake: an experimental study exploring lake compartments. Sci Total Environ. 2022;803:150049. https://doi.org/10.1016/j.scitotenv.2021.150049 .
Hanson PC, Hamilton DP, Stanley EH, Preston N, Langman OC, Kara EL. Fate of allochthonous dissolved organic carbon in lakes: a quantitative approach. PLoS ONE. 2011;6(7). https://doi.org/10.1371/journal.pone.0021884 .
Seymour JR, Amin SA, Raina J-B, Stocker R. Zooming in on the phycosphere: the ecological interface for phytoplankton-bacteria relationships. Nat Microbiol. 2017;2(7):17065. https://doi.org/10.1038/nmicrobiol.2017.65 .
Zhao L, Lin LZ, Zeng Y, Teng WK, Chen MY, Brand JJ, et al. The facilitating role of phycospheric heterotrophic bacteria in cyanobacterial phosphonate availability and Microcystis bloom maintenance. Microbiome. 2023;11(1):142. https://doi.org/10.1186/s40168-023-01582-2 .
Xie P. Biological mechanisms driving the seasonal changes in the internal loading of phosphorus in shallow lakes. Sci China Ser D-Earth Sci. 2006;49:14–27. https://doi.org/10.1007/s11430-006-8102-z .
Xue J, Yao X, Zhao Z, He C, Shi Q, Zhang L. Internal loop sustains cyanobacterial blooms in eutrophic lakes: evidence from organic nitrogen and ammonium regeneration. Water Res. 2021;206. https://doi.org/10.1016/j.watres.2021.117724 .
Paver SF, Hayek KR, Gano KA, Fagen JR, Brown CT, Davis-Richardson AG, et al. Interactions between specific phytoplankton and bacteria affect lake bacterial community succession. Environ Microbiol. 2013;15(9):2489–504. https://doi.org/10.1111/1462-2920.12131 .
Miki T, Jacquet S. Complex interactions in the microbial world: underexplored key links between viruses, bacteria and protozoan grazers in aquatic environments. Aquat Microb Ecol. 2008;51(2):195–208.
Chow C-ET, Kim DY, Sachdeva R, Caron DA, Fuhrman JA. Top-down controls on bacterial community structure: microbial network analysis of bacteria, T4-like viruses and protists. ISME J. 2014;8(4):816–29. https://doi.org/10.1038/ismej.2013.199 .
Pradeep Ram AS, Keshri J, Sime-Ngando T. Distribution patterns of bacterial communities and their potential link to variable viral lysis in temperate freshwater reservoirs. Aquat Sci. 2019;81(4):72. https://doi.org/10.1007/s00027-019-0669-5 .
Landi P, Minoarivelo HO, Brannstrom A, Hui C, Dieckmann U. Complexity and stability of ecological networks: a review of the theory. Popul Ecol. 2018;60(4):319–45. https://doi.org/10.1007/s10144-018-0628-3 .
Pan RK, Sinha S. Modular networks emerge from multiconstraint optimization. Phys Rev E. 2007;76(4). https://doi.org/10.1103/PhysRevE.76.045103 .
Alcantara JM, Rey PJ. Linking topological structure and dynamics in ecological networks. Am Nat. 2012;180(2):186–99. https://doi.org/10.1086/666651 .
Dai W, Zhang J, Tu Q, Deng Y, Qiu Q, Xiong J. Bacterioplankton assembly and interspecies interaction indicating increasing coastal eutrophication. Chemosphere. 2017;177:317–25. https://doi.org/10.1016/j.chemosphere.2017.03.034 .
Deng Y, Jiang YH, Yang YF, He ZL, Luo F, Zhou JZ. Molecular ecological network analyses. BMC Bioinformatics. 2012;13:113. https://doi.org/10.1186/1471-2105-13-113 .
Liu S, Ji Z, PU F, Liu Y, Zhou S, Zhai J. On phytoplankton community composition structure and biological assessment of water trophic state in Xingyun Lake. J Saf Environ. 2019;19(4):1439–47.
Download references
Acknowledgements
We express our gratitude to Wei Tian, Jingchen Xue, Meng Qu, Keqiang Shao, and Dong Li for their valuable assistance in sample collection and laboratory measurements. Additionally, we appreciate the editors and anonymous reviewers for their constructive suggestions and comments, which have greatly contributed to the refinement of this work.
This study received financial support from the National Natural Science Foundation of China (grant numbers: 41971062, 42371065) and West Anhui University through the “Scientific Research Start-up Funds for High-level Talents” (WGKQ2022032).
Author information
Authors and affiliations.
College of Biology and Pharmaceutical Engineering, West Anhui University, Lu’an, 237012, China
Guijuan Xie
Taihu Laboratory for Lake Ecosystem Research, State Key Laboratory of Lake Science and Environment, Nanjing Institute of Geography and Limnology, Chinese Academy of Sciences, Nanjing, 210008, China
Guijuan Xie, Yuqing Zhang, Yi Gong, Wenlei Luo & Xiangming Tang
The Third Construction Company of CCCC second Harbor Engineering Co., Ltd, Zhenjiang, 212000, China
Yuqing Zhang
The Fuxianhu Station of Plateau Deep Lake Field Scientific Observation and Research, Yunnan, 653100, Yuxi, China
You can also search for this author in PubMed Google Scholar
Contributions
G.X. and X.T. conceived and conducted the experiment. G.X., Y.Z., Y.G., and W.L. analyzed the results, and wrote the article; X.T. revised the manuscript. All authors reviewed the manuscript. No conflict of interest exists in the submission of this manuscript, and the final manuscript is approved by all authors for publication.
Corresponding author
Correspondence to Xiangming Tang .
Ethics declarations
Ethics approval and consent to participate.
Not applicable.
Consent for publication
Competing interests.
The authors declare no competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1
Supplementary material 2, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/ .
Reprints and permissions
About this article
Cite this article.
Xie, G., Zhang, Y., Gong, Y. et al. Extreme trophic tales: deciphering bacterial diversity and potential functions in oligotrophic and hypereutrophic lakes. BMC Microbiol 24 , 348 (2024). https://doi.org/10.1186/s12866-024-03488-x
Download citation
Received : 18 February 2024
Accepted : 02 September 2024
Published : 14 September 2024
DOI : https://doi.org/10.1186/s12866-024-03488-x
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Lake Fuxian
- Lake Xingyun
- Oligotrophic
- Hypereutrophic
- Bacterial diversity
- Network complexity and stability
BMC Microbiology
ISSN: 1471-2180
- General enquiries: [email protected]
Causal relationship between Lipdome and Chronic Obstructive Pulmonary Disease and Asthma: Mendelian randomization
- Original Article
- Open access
- Published: 25 September 2024
- Volume 14 , article number 249 , ( 2024 )
Cite this article
You have full access to this open access article

- Qiong Wu ORCID: orcid.org/0009-0008-4425-2062 1 na1 ,
- Jingmin Fu ORCID: orcid.org/0009-0003-8790-6349 2 na1 ,
- Cheng Zhang ORCID: orcid.org/0009-0000-6121-7754 3 ,
- Zhuolin Liu ORCID: orcid.org/0009-0009-8839-5396 3 ,
- Jianing Shi ORCID: orcid.org/0009-0004-3489-7933 2 ,
- Zhiying Feng ORCID: orcid.org/0009-0000-9125-4687 2 ,
- Kangyu Wang ORCID: orcid.org/0009-0001-9954-9890 2 &
- Ling Li ORCID: orcid.org/0000-0002-9900-3695 3
89 Accesses
Explore all metrics
Genetic risk significantly influence susceptibility and heterogeneity of chronic obstructive pulmonary disease (COPD) and asthma, and increasing evidence suggests their close association with lipdome. However, their causal relationship remains unclear. In this study, we conducted a two-sample MR (Mendelian randomization) analysis using publicly available large-scale genome-wide association studies (GWAS) data to evaluate the causal impact of lipdome on COPD and asthma. The inverse variance weighted (IVW) method served as the primary analysis method, and multiple sensitivity and heterogeneity tests were performed to assess the reliability of the results. Finally, a Meta-analysis was conducted on lipdome with significant causal relationships to validate the robustness of the results. Our findings suggest that Sterol ester (27:1/18:2), Phosphatidylcholine (15:0_18:2), (16:0_18:2), (16:0_20:2), (17:0_18:2), (18:1_18:1), (18:1_18:2), (18:1_20:2), Triacylglycerol (54:3), and (56:4) levels are protective factors for COPD, while levels of Phosphatidylcholine (16:0_22:5), (18:0_20:4), and (O-16:0_20:4) are risk factors for COPD. Meta-analysis of lipids causally related to COPD also indicates significant results. Phosphatidylcholine (16:0_20:4), (16:0_22:5), and (18:0_20:4) levels are risk factors for asthma, while Phosphatidylcholine (18:1_18:2), (18:1_20:2), and Sphingomyelin (d38:1) levels are protective factors for asthma. However, the lack of statistical significance in the Meta-analysis may be due to heterogeneity in research methods and data statistics. This study indicates that 4 lipdome species have significant correlations with COPD and asthma. Phosphatidylcholine (18:1_18:2) and (18:1_20:2) are protective factors, while Phosphatidylcholine (16:0_22:5) and (18:0_20:4) are risk factors. Additionally, due to differences in molecular subtypes, phosphatidylcholine, sterol ester, and triacylglycerol exhibit differential effects on the diseases.
Similar content being viewed by others
Exploring the causal effect between lipid-modifying drugs and idiopathic pulmonary fibrosis: a drug-target Mendelian randomization study

Genetic Association of Circulating Adipokines with Risk of Idiopathic Pulmonary Fibrosis: A Two-Sample Mendelian Randomization Study
Repurposing lipid-lowering drugs on asthma and lung function: evidence from a genetic association analysis
Avoid common mistakes on your manuscript.
Introduction
Chronic respiratory diseases (CRD) encompass diseases of the airways and other structures of the lungs, including COPD, asthma, interstitial lung disease, occupational lung diseases, and lung nodules (Gould et al. 2023 ). Globally, CRD accounts for a significant number of deaths, disability-adjusted life years (DALYs), incidences, and prevalence, making it a leading cause of disability and death worldwide. The healthcare costs associated with respiratory diseases continue to rise, imposing a heavy economic burden on nations and individuals (GBD Chronic Respiratory Disease Collaborators 2020 ; GBD 2019 Chronic Respiratory Diseases Collaborators 2023 ).
COPD and asthma are complex inflammatory diseases of the airways and represent the two main diseases within CRD. COPD is the leading cause of death among CRD patients, while asthma has the highest prevalence within CRD (GBD Chronic Respiratory Disease Collaborators 2020 ). Both asthma and COPD are heterogeneous diseases with a range of underlying mechanisms, characterized by airflow limitation (Dasgupta et al. 2023 ). Moreover, the pathological mechanisms of COPD and asthma are closely related to airway epithelial cell reprogramming and immune cell-mediated inflammatory responses (Miller et al. 2021 ; Christenson et al. 2022 ). However, COPD is often undiagnosed and untreated in its early stages, only receiving attention when symptoms become severe. Simultaneously, asthma has become the most common chronic disease in pediatrics, affecting adolescents and adults (Bitsko et al. 2014 ). The phenotypes and underlying mechanisms of COPD and asthma are not yet fully understood. It is now widely believed that asthma susceptibility has a strong genetic component (Ntontsi et al. 2021 ), and genetic risk plays a crucial role in the susceptibility and heterogeneity of COPD (Christenson et al. 2022 ).
Lipidome refers to the entire collection of chemically distinct lipid species in cells, organs, or biological systems (Kishimoto et al. 2001 ). Lipids are important cellular components that play critical roles in cell structure formation, cellular signal transduction, and bioenergetics (Hornburg et al. 2023 ). Increasing evidence suggests that dysregulation of lipid metabolism is closely associated with the pathogenesis of COPD and asthma, as it influences the occurrence and progression of these diseases (Kotlyarov and Bulgakov 2021 ; Loureiro et al. 2016 ). For example, in obese populations with high levels of triglycerides and cholesterol, poorer outcomes in COPD have been observed (Lambert et al. 2017 ). Cholesterol overload has been linked to COPD and airway epithelium-driven inflammation (Li et al. 2022 ). In asthma allergic models, sphingolipid mediators, such as sphingosine-1-phosphate and ceramide, have been shown to be important signaling molecules in airway hyperresponsiveness, mast cell activation, and inflammation (Ono et al. 2015 ).These different types of lipids play various roles in organisms, and among all lipid categories, mammalian cells may have thousands of individual lipid species, collectively referred to as the lipidome (Raghu 2020 ). Since the lipidome is composed of one or more concentric phospholipid bilayers surrounding a water core to form spherical vesicles, it has been widely used in the field of nanomedicine. Specific lipids are used as drug delivery systems, enhancing the precise targeted treatment of drugs (Cao et al. 2024 ). The lipidome has also been shown to be a new participant in the pathophysiology of COPD and asthma. Pulmonary surfactant is distributed on the surface of the alveolar fluid molecular layer in the lungs, with the main component being saturated phosphatidylcholine, such as phosphatidylcholine 16:0/16:0, phosphatidylcholine 16:0/14:0, and phosphatidylcholine 16:0/16:1. It can maintain the relative stability of the size of the alveolar volume (Eggers et al. 2017 ). Therefore, further research on the role of the lipidome in COPD and asthma may provide new insights for the comprehensive improvement of COPD and asthma management (Donnelly 2014 ; Ravi et al. 2021 ).
In the past few decades, genome-wide association studies (GWAS) have become an important research method in genetics. Mendelian randomization (MR) is a statistical method used to infer causal relationships, using summary data from GWAS to select instrumental variables (IVs) in the form of single nucleotide polymorphisms (SNPs) that meet certain conditions. These IVs are then used to explore causal relationships between exposures and outcomes (Birney 2022 ). Due to the random allocation of genetic variation during conception, MR reduces confounding factors and reverse causation compared to randomized controlled trials, saving a significant amount of time and effort (Davey Smith and Hemani 2014 ). In recent years, MR has been widely used to explore associations between various cellular molecules and a wide range of diseases. However, there is little evidence investigating the causal relationships between the lipidome and COPD or asthma. Therefore, this study aims to use large-scale GWAS data to identify potential causal relationships between the lipidome and COPD and asthma, providing new insights for the diagnosis and treatment of these conditions.
The emergence of high-sensitivity mass spectrometry has enabled people to determine an increasing number of lipid species, making lipid nomenclature both easier to manage and more descriptive (Kopczynski et al. 2020 ). This study provides a detailed classification of lipid species, such as phosphatidylcholine belonging to the glycerophospholipid class, and phosphatidylcholine (15:0_18:2) belonging to the molecular subtype. ‘15’ represents the number of carbon atoms, '0' indicates the number of double bonds, which are used to determine the length and saturation of fatty acyl chains (FA). (15:0_18:2) indicates that this phosphatidylcholine has two FA chains, with the second chain having 18 carbon atoms and 2 double bonds. Even within the same class of phospholipids, the structure determines the more specific functions of lipid bodies. Therefore, the results of this study further refine the analysis to the subtype level.
Materials and methods
Study design.
In this study, we utilized significantly associated SNPs from the GWAS data of the lipidome as IVs, with COPD and asthma as outcomes. We conducted a two-sample MR analysis using summary statistics from GWAS to assess the causal relationships between the lipidome and COPD and asthma. To minimize the impact of confounding factors and avoid reverse causality, the standard MR analysis needs to meet the following three assumptions (Davies et al. 2018 ): (1) the IV is associated with the exposure factor; (2) the IV is independent and unrelated to any confounding factors; (3) the IV only affects the outcome through the pathway of the exposure factor. In addition, to further validate the results of the MR analysis, we performed a Mendelian randomization pleiotropy residual sum and Meta-analysis on the results with significant causal relationships.
Data sources
The summary data for the lipidome were obtained from a recent publication on whole-genome association analysis of human plasma lipidomes. The study included 7174 individuals from the Finnish population and detected 179 lipid species belonging to 13 lipid categories using shotgun lipidomics. The four major lipid categories covered were glycerolipids, glycerophospholipids, sphingolipids, and sterols. The study conducted univariate and multivariate genome-wide analyses and identified 495 genome-wide significant loci, including 56 genetic loci (8 of which were newly identified) (Ottensmann et al. 2023 ).
The statistical data for COPD and asthma were retrieved from the publicly available GWAS database ( https://gwas.mrcieu.ac.uk/ ). The COPD dataset included a total of 468,475 individuals, with 454,945 healthy controls and 13,530 COPD-positive patients. The number of SNPs in the dataset was 24,180,654. The asthma dataset included a total of 484,598 individuals, with 428,511 healthy controls and 56,087 asthma-positive patients. The number of SNPs in the dataset was 9,587,836. (COPD GWAS ID: ebi-a-GCST90018807; Asthma GWAS ID: ebi-a-GCST90038616).
Selection of instrumental variables
To identify SNPs with significant correlations, we set the significance threshold at P < 1.0 × 10 –5 . However, this led to a large number of results, so we further raised the threshold. To identify strongly positive SNPs, the final threshold was set at P < 1.0 × 10 –8 (Yu et al. 2023 ). To eliminate linkage disequilibrium, we set the clustering window at 10,000 kb and an r 2 threshold at 0.001 to cluster SNPs and obtain independent loci. In addition, we calculated the F -statistic for each SNP to measure the strength of each SNP's instrumentality, with SNPs having an F < 10 considered weak instrumental variables (Li et al. 2023 ).
MR analysis
First, we used the IVW as the primary MR method to assess the relationship between the lipidome and COPD and asthma. This method combines the Wald values for each SNP to estimate the effect, which is equivalent to weighted linear regression of the associations between instrumental variables, providing estimates unaffected by horizontal pleiotropy (Burgess et al. 2013 ). To further screen for significant results and account for the influence of pleiotropy, we used MR-Egger regression, weighted median, simple mode, and weighted mode as supplementary methods. The weighted median method can provide consistent estimates even with up to 50% invalid SNPs (Burgess et al. 2017 ), and MR-Egger can generate unbiased estimates of causal effects even when all instrumental SNPs exhibit pleiotropy (Bowden et al. 2015 ). Additionally, we used MR-Egger intercept to determine the presence of horizontal pleiotropy ( P < 0.05 considered significant). Furthermore, we conducted heterogeneity analysis on the results, with P < 0.05 considered evidence of heterogeneity. Finally, we performed leave-one-out analysis to ensure that the MR results were not unduly influenced by individual SNPs.
Meta-analysis of MR results
Meta-analysis is a statistical method used to compare and synthesize the results of studies on the same scientific question. By integrating all relevant studies, it provides robustness in interpreting the results. After the Meta-analysis results of MR analysis are subjected to tests for heterogeneity and multiple effects, lipid nanoparticles with more significant results are selected for Meta-analysis with relevant diseases. Cochran’s Q test is used to evaluate the heterogeneity of the results, and P < 0.05 indicates significant heterogeneity between the studies. In Meta-analysis, a random-effects model is chosen if there is significant heterogeneity ( P < 0.05), otherwise ( P > 0.05), a fixed-effects model is selected.
Using IVW analysis as the main method, our study identified a total of 19 lipids with potential causal relationships with COPD, with four lipids associated with increased COPD risk and 15 lipids associated with decreased COPD risk (Fig. 1 ). In the results of asthma, a total of 26 lipids were found to have potential causal relationships, with 12 lipids associated with increased asthma risk and 14 lipids associated with decreased asthma risk (Fig. 2 ).
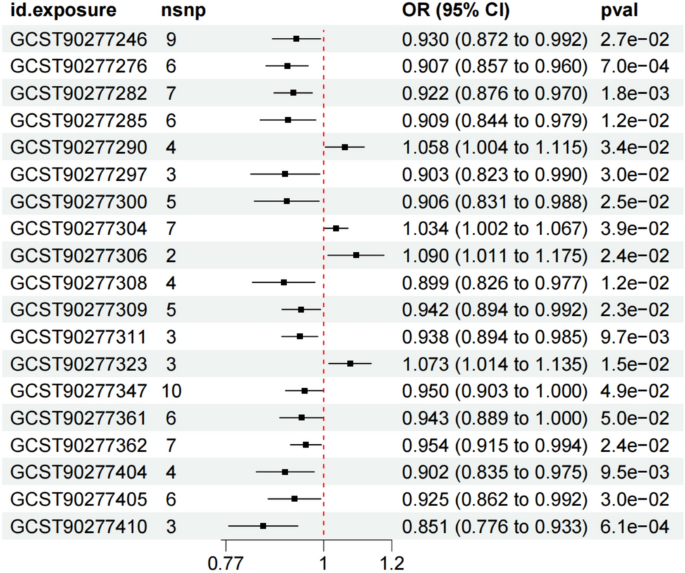
The causal relationships between 19 lipdome and COPD identified through IVW analysis, including specific exposure ID, the number of SNPs, OR value, 95% confidence interval, and p-value
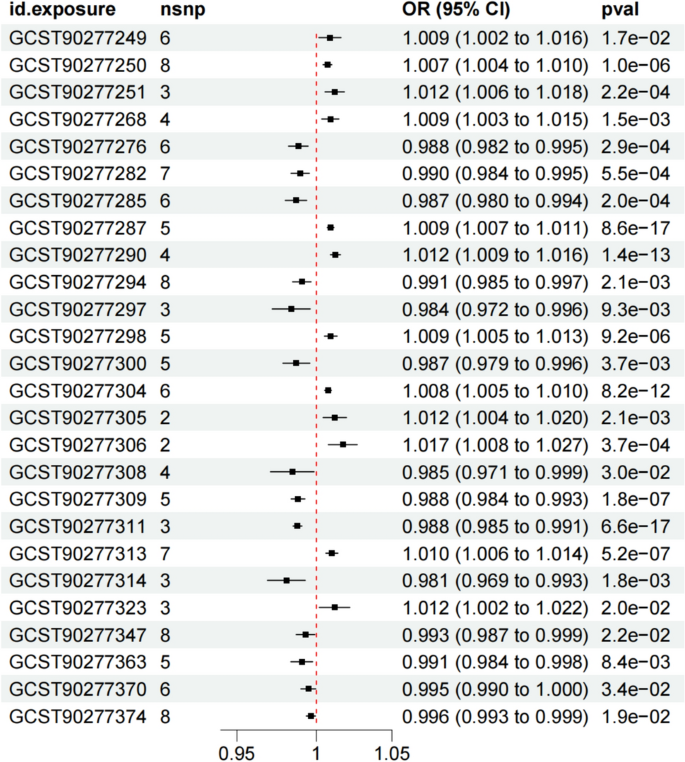
The causal relationships between 26 lipdome and asthma identified through IVW analysis, including specific exposure ID, the number of SNPs, OR value, 95% confidence interval, and p-value
In the sensitivity analysis of lipids and COPD (Figs. 3 , 4 , and Table 1 ), besides the IVW method, Sterol ester (27:1/18:2) levels (OR 0.930, 95% CI 0.872–0.992, P ivw = 2.7e−02) also had a P -value less than 0.05 in the Weighted Median and Weighted Mode analyses, indicating a significant causal relationship with COPD and a lower risk of COPD (Fig. 5 ). In addition, 12 lipids showed significant causal relationships with COPD risk in both IVW and Weighted Median analyses, with P < 0.05, and 9 lipids were associated with a decreased risk of COPD, including: Phosphatidylcholine (15:0_18:2) levels (OR 0.907, 95% CI 0.857–0.960, P ivw = 7.0e−04), Phosphatidylcholine (16:0_18:2) levels (OR 0.922, 95% CI 0.876–0.970, P ivw = 1.8e−03), Phosphatidylcholine (16:0_20:2) levels (OR 0.909, 95% CI 0.844–0.979, P ivw = 1.2e−02), Phosphatidylcholine (17:0_18:2) levels (OR 0.903, 95% CI 0.823–0.990, P ivw = 3.0e−02), Phosphatidylcholine (18:1_18:1) levels (OR 0.899, 95% CI 0.826–0.977, P ivw = 1.2e−02), Phosphatidylcholine (18:1_18:2) levels (OR 0.942, 95% CI 0.894–0.992, P ivw = 2.3e−02), Phosphatidylcholine (18:1_20:2) levels (OR 0.938, 95% CI 0.894–0.985, P ivw = 9.7e−03), Triacylglycerol (54:3) levels (OR 0.902, 95% CI 0.835–0.975, P ivw = 9.5e−03), and Triacylglycerol (56:4) levels (OR 0.851, 95% CI 0.776–0.933, P ivw = 6.1e−04). Three lipids were associated with increased COPD risk, including: Phosphatidylcholine (16:0_22:5) levels (OR 1.058, 95% CI 1.004–1.115, P ivw = 3.4e−02), Phosphatidylcholine (18:0_20:4) levels (OR 1.304, 95% CI 1.002–1.067, P ivw = 3.9e−02), and Phosphatidylcholine (O-16:0_20:4) levels (OR 1.073, 95% CI 1.014–1.135, P ivw = 1.5e−02). The MR-Egger intercepts of all lipids were P > 0.05, indicating no horizontal pleiotropy. Phosphatidylcholine (18:0_22:5) levels could not be calculated for other analysis methods due to insufficient SNPs, and the other 18 lipids showed no heterogeneity. In the leave-one-out analysis, no significant abnormal SNPs were found for all lipids.
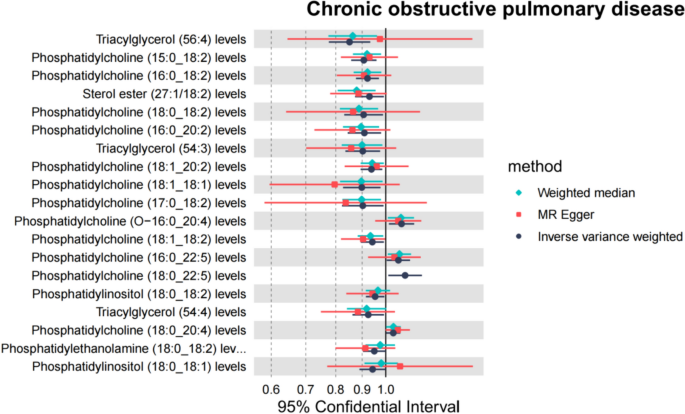
The sensitivity analysis results of lipdome and COPD include Weighted median, MR Egger and IVW
Circular plot for the sensitivity analysis of lipdome and COPD, lipdome that have significance ( Pivw < 0.05) have been marked
Results of the five MR analyses for Sterol ester (27:1/18:2) levels against COPD
In the results of sensitivity analysis for lipids and asthma (Figs. 6 , 7 , and Table 2 ), besides insthe IVW method, 12 lipids showed a P < 0.05 in the MR Egger, weighted median, and weighted mode analyses, 9 lipids showed a P < 0.05 in the weighted median and weighted mode analyses, and 2 lipids showed a P < 0.05 in the weighted median analysis. However, among these lipids, 17 lipids had an MR-Egger intercept P < 0.05, indicating horizontal pleiotropy, 4 lipids showed heterogeneity, and 2 lipids did not have enough SNPs. Finally, we identified causal relationships between 6 lipids and asthma, with 4 lipids (based on 4 MR analyses with P < 0.05) showing a significant causal relationship with asthma (Fig. 8 ). These lipids include: Phosphatidylcholine (16:0_20:4) levels (OR 1.009, 95% CI 1.007–1.011, P ivw = 8.6e−17), Phosphatidylcholine (16:0_22:5) levels (OR 1.012, 95% CI 1.009–1.016, P ivw = 1.4e−13), and Phosphatidylcholine (18:0_20:4) levels (OR 1.008, 95% CI 1.005–1.010, P ivw = 8.2e−12), which are associated with an increased risk of asthma, and Phosphatidylcholine (18:1_18:2) levels (OR 0.988, 95% CI 0.984–0.993, P ivw = 1.8e−07), which is associated with a decreased risk of asthma. One lipid (based on 3 MR analyses with P < 0.05) showed a significant causal relationship with asthma and a decreased risk of asthma: Phosphatidylcholine (18:1_20:2) levels (OR 0.988, 95% CI 0.985–0.991, P ivw = 6.6e−17). One lipid (based on 2 MR analyses with P < 0.05) showed a relatively significant causal relationship with asthma and a decreased risk of asthma: Sphingomyelin (d38:1) levels (OR 0.996, 95% CI 0.993–0.999, Pivw = 1.9e−02). In the leave-one-out analysis, we found that three SNPs might have a significant impact on the results, with SNP rs174581 having a large impact on Sterol ester (27:1/20:5) levels, SNP rs174533 having a large impact on Phosphatidylcholine (17:0_20:4) levels, and SNP rs174568 having a large impact on Phosphatidylcholine (O-16:0_20:4) levels (Fig. 9 ).
The sensitivity analysis results of lipdome and asthma include Weighted median, MR Egger and IVW
Circular plot for the sensitivity analysis of lipids and asthma, lipdome that have significance ( Pivw < 0.05) have been marked
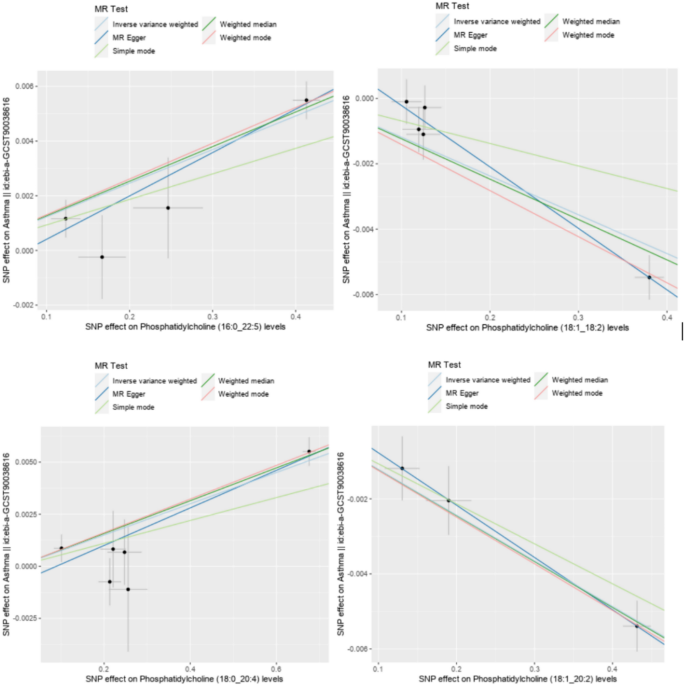
MR analysis of four lipdome with significant causal associations with asthma and COPD
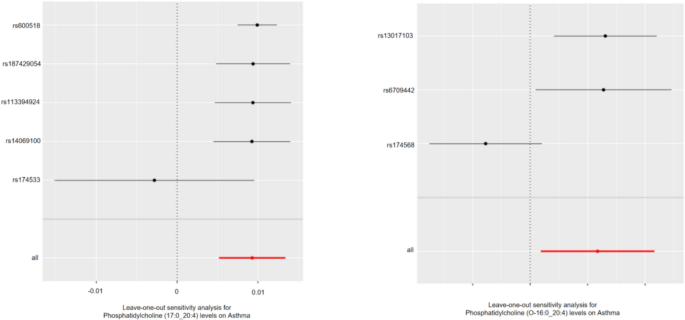
Leave-one-out analysis reveals two individual SNPs from lipids that have an impact on the outcome, SNP rs174533 having a large impact on Phosphatidylcholine (17:0_20:4) levels, and SNP rs174568 having a large impact on Phosphatidylcholine (O-16:0_20:4) levels
Meta-analysis was conducted on 13 lipid components that have a causal relationship with COPD (Fig. 10 ). According to the results of Cochran's Q test, due to the heterogeneity ( P < 0.0001), a random-effects model was selected, and the meta-analysis showed a significant result ( P < 0.05), indicating a potential correlation between lipids and COPD. Similarly, a causal analysis was conducted on 6 lipid components that have a causal relationship with asthma (Fig. 11 ). The heterogeneity ( P < 0.0001) also led to the selection of a random-effects model. However, the Meta-analysis with P = 0.948 indicated no statistical significance.
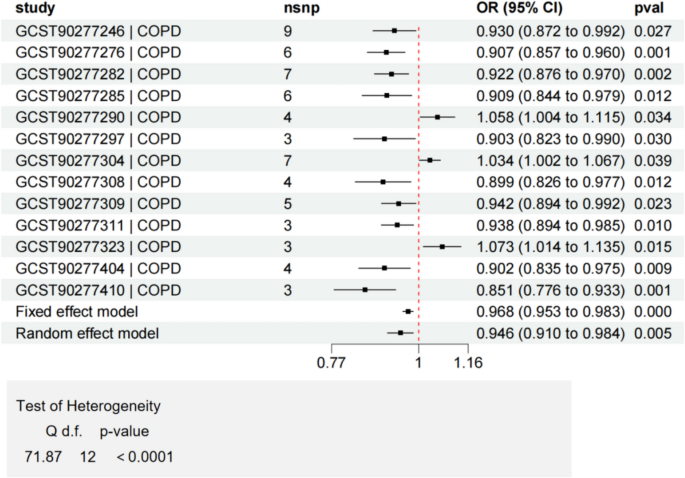
Meta analysis of lipids with significant causal associations with COPD, heterogeneity test ( P < 0.0001)
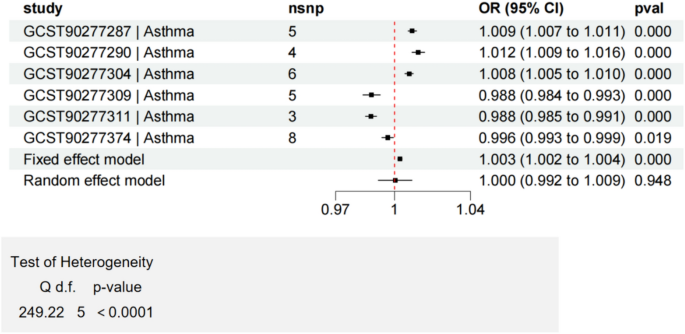
Meta analysis of lipids with significant causal associations with asthma, heterogeneity test ( P < 0.0001)
In this study, we conducted two-sample MR analysis using large-scale GWAS data. Our results indicate that there is a causal relationship between 13 lipid components and COPD, In this study, large-scale GWAS data were used for two-sample Mendelian randomization (MR) analysis. Our results indicate causal relationships between 13 lipid species and COPD. Sterol ester (27:1/18:2), Phosphatidylcholine (15:0_18:2), Phosphatidylcholine (16:0_18:2), Phosphatidylcholine (16:0_20:2), Phosphatidylcholine (17:0_18:2), Phosphatidylcholine (18:1_18:1), Phosphatidylcholine (18:1_18:2), Phosphatidylcholine (18:1_20:2), Triacylglycerol (54:3), and Triacylglycerol (56:4) levels are protective factors for COPD, while Phosphatidylcholine (16:0_22:5), Phosphatidylcholine (18:0_20:4), and Phosphatidylcholine (O-16:0_20:4) levels are risk factors for COPD. Meta analysis for lipid species causally associated with COPD also yielded significant results. In the MR analysis of asthma, we identified 6 lipid species that are causally related to asthma. Phosphatidylcholine (16:0_20:4), Phosphatidylcholine (16:0_22:5), and Phosphatidylcholine (18:0_20:4) levels are risk factors for asthma, while Phosphatidylcholine (18:1_18:2), Phosphatidylcholine (18:1_20:2), and Sphingomyelin (d38:1) levels are protective factors for asthma. However, the Meta analysis did not show statistical significance, which could be attributed to methodological and data heterogeneity.
Numerous previous metabolomics and lipidomics studies have indicated a correlation between lipids and chronic lung diseases. Some lipid metabolites may serve as potential biomarkers for screening, diagnosing, and treating COPD and asthma. In our study, we identified Sterol ester as the most relevant lipid component associated with COPD. As indicated in the study by Luo et al., the elevated cholesterol levels in peripheral blood are positively correlated with disease severity in patients with COPD or smokers (Luo et al. 2020 ). Additionally, Reed et al. found that severe COPD has been associated with increased levels of high-density lipoprotein cholesterol (Reed et al. 2011 ). Moreover, Sugiura et al. found that the content of cholesterol 25-hydroxycholesterol in lung tissue of COPD patients is significantly higher compared to normal individuals (Sugiura et al. 2012 ). Angelidis et al. discovered that cholesterol-related metabolism in lung epithelial cells and lipid fibroblasts is considered a potential marker for lung aging (Angelidis et al. 2019 ). However, our results indicate an inverse relationship between Sterol ester and the risk of developing COPD, which is inconsistent with the majority of previous studies, this may be related to the molecular subtype of Sterol ester. Among the lipid components associated with COPD, most of them are Phosphatidylcholine, but the differences in molecular subtypes (carbon atom number, double bond number, different positions, etc.) led to different effects of Phosphatidylcholine on COPD, Additionally, some were Triacylglycerol, indicating the need for further research to confirm the molecular subtypes of these lipid species. A study identified over 40 lipid compounds in plasma samples from COPD patients, and Phosphatidylcholine 34:3 and Triacylglycerol 52:3 were identified as potential biomarkers associated with disease severity and oxidative status in COPD (Angelidis et al. 2019 ). Additionally, the reduction of phospholipid and glycerol phospholipid is related to lung surfactant damage (Angelidis et al. 2019 ). Abnormal glycerophospholipid metabolism may be associated with the pathogenesis of non-eosinophilic subtype COPD, and the expression of lysophosphatidylcholine 18:3, lysophosphatidylethanolamine 16:1, and phosphatidylinositol 32:1 were significantly reduced in the acute exacerbation and recovery period of COPD compared to stable period (Gai et al. 2021 ). In a clinical study on stable chronic bronchitis in adults, inhalation of phosphatidylcholine (PC) 32:0 (the most abundant surfactant phospholipid) improved lung function in patients (Anzueto et al. 1997 ). A Meta-analysis showed that in the subgroup analysis of stable COPD patients who did not receive lipid-lowering therapy, their triglyceride levels were higher than those of healthy individuals (Xuan et al. 2018 ).
In asthma-related lipdome, the majority are Phosphatidylcholine, and there is also a type called sphingomyelin. Previous lipidomics studies have shown that the fatty acid metabolism in the bronchial epithelial cells of asthma patients is altered, leading to elevated levels of certain lipid species [phosphatidylcholine, lyso-phosphatidylcholine, and diacylglycerol phosphate], which are associated with the pathophysiology of asthma (Ravi et al. 2021 ). Similarly, metabolomics studies have found that elevated levels of various phosphatidylcholine and decreased levels of various lyso-phosphatidylcholine are associated with asthma (Ried et al. 2013 ). Furthermore, lipidomics studies have also discovered abnormal lipid metabolism in asthma patients, with significant changes in 10 lipid species in plasma, which are associated with the severity of asthma and IgE levels (Jiang et al. 2021 ). After inhalation of allergens, asthma patients have increased levels of oxidized phosphatidylcholine in the stimulated airways. Oxidized phosphatidylcholine may promote the pathobiology of asthma through inducing pro-inflammatory phenotypes and airway smooth muscle contraction, making it a potential new therapeutic target for oxidative stress-related pathobiology in asthma (Pascoe et al. 2021 ). Additionally, in asthma patients, the levels of bioactive hemolytic phosphatidylcholines 16:0 and 18:0 are significantly elevated in the lungs. The increase in hemolytic phosphatidylcholine content may be a potential key lipid mediator underlying the occurrence and progression of airway epithelial injury in asthma (Yoder et al. 2014 ). In a clinical study, compared to the healthy control group, asthma patients showed significantly decreased levels of sphingomyelin species (sphingomyelin 34:2, sphingomyelin 38:1, and sphingomyelin 40:1). In non-eosinophilic asthmatics, the serum levels of sphingomyelin were significantly reduced, which is consistent with our results (Guo et al. 2021 ). Sphingolipid metabolites are important mediators for obesity-related asthma, pediatric asthma, and exacerbation of respiratory system disease-induced airway inflammation (Guo et al. 2021 ). More importantly, sphingolipids have been shown to play a role in the pathogenesis of bronchopulmonary dysplasia. Interventions that interfere with sphingolipid metabolism may be a novel strategy to prevent and repair lung diseases, which could potentially reduce the incidence and mortality of severe lung diseases (Tibboel et al. 2014 ).
Overall, phosphatidylcholine or its subtypes show significant potential research value in COPD and asthma. More importantly, among the four lipdome identified in this study with significant implications for COPD and asthma, their impact on disease trends is consistent. Phosphatidylcholine (18:1_18:2) and Phosphatidylcholine (18:1_20:2) are protective factors, while Phosphatidylcholine (16:0_22:5) and Phosphatidylcholine (18:0_20:4) are risk factors. In recent years, the overlap in pathogenic mechanisms (such as airway inflammation), treatment methods (such as steroids), and disease gene networks between asthma and COPD suggests a high degree of similarity in the pathobiology of these two diseases (Hizawa 2024 ). Although phosphatidylcholine is the main component of pulmonary surfactant, research has also found significant amounts of phosphatidylglycerol [36:1], phosphatidylglycerol [34:1], and phosphatidylglycerol [34:2] in non-tumor lung tissue, all of which are components of pulmonary surfactants (Eggers et al. 2017 ). Therefore, the lipdome identified in this study are of great significance for the diagnosis and treatment of COPD and asthma.
This study has several strengths. The main advantage is that it is the first to use MR analysis to validate the causal relationship between lipdome and COPD and asthma, which can reduce confounding factors and reverse causality biases. Asthma is a genetically predisposed disease, and COPD has been shown to be associated with genetic risk. This study validates the relationship between lipdome and these diseases through large-scale GWAS data. MR, as an important method in genetic research, has great epidemiological significance. This study also further complements the previous lipidomics views on lung-related diseases and provides some ideas for clinical diagnosis and treatment. However, our study also has some limitations. Firstly, this study focuses on the broad scope of research on COPD and asthma, without staging COPD or classifying asthma. Secondly, all the GWAS data are derived from European populations, and whether the results are applicable to other populations needs further verification. Thirdly, in the Mendelian randomization analysis of lipdome related to asthma, the p-value we obtained did not reach statistical significance and contradicted some previous study conclusions, which may affect the accuracy of the research. Finally, although we rigorously screened the instrumental variables (significance threshold P < 1.0 × 10 –8 ), our conclusions still need to be verified by randomized controlled experiments with high levels of evidence.
In conclusion, this study utilized publicly available large-scale GWAS data for two-sample MR analysis to assess the causal effects of lipid species on COPD and asthma. 4 lipdome species showed significant correlations with COPD and asthma. Phosphatidylcholine (18:1_18:2) and Phosphatidylcholine (18:1_20:2) are protective factors, while Phosphatidylcholine (16:0_22:5) and Phosphatidylcholine (18:0_20:4) are risk factors. Due to differences in molecular subtypes, certain phosphatidylcholine, sterol ester, and triacylglycerol also exhibit differential effects on the diseases. These complex molecular mechanisms provide further insights into the application of lipidomics in COPD and asthma.
Data availability
This study used publicly available GWAS data ( https://gwas.mrcieu.ac.uk/ ), and the relevant results of this study have been included in the supplementary files.
Angelidis I, Simon LM, Fernandez IE, Strunz M, Mayr CH, Greiffo FR, Tsitsiridis G, Ansari M, Graf E, Strom TM, Nagendran M, Desai T, Eickelberg O, Mann M, Theis FJ, Schiller HB (2019) An atlas of the aging lung mapped by single cell transcriptomics and deep tissue proteomics. Nat Commun 10(1):963. https://doi.org/10.1038/s41467-019-08831-9
Article CAS PubMed PubMed Central Google Scholar
Anzueto A, Jubran A, Ohar JA, Piquette CA, Rennard SI, Colice G, Pattishall EN, Barrett J, Engle M, Perret KA, Rubin BK (1997) Effects of aerosolized surfactant in patients with stable chronic bronchitis: a prospective randomized controlled trial. JAMA 278(17):1426–1431
Article CAS PubMed Google Scholar
Birney E (2022) Mendelian randomization. Cold Spring Harb Perspect Med 12(4):a041302. https://doi.org/10.1101/cshperspect.a041302
Article PubMed PubMed Central Google Scholar
Bitsko MJ, Everhart RS, Rubin BK (2014) The adolescent with asthma. Paediatr Respir Rev 15(2):146–153. https://doi.org/10.1016/j.prrv.2013.07.003
Article PubMed Google Scholar
Bowden J, Davey Smith G, Burgess S (2015) Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 44(2):512–525. https://doi.org/10.1093/ije/dyv080
Burgess S, Butterworth A, Thompson SG (2013) Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol 37(7):658–665. https://doi.org/10.1002/gepi.21758
Burgess S, Bowden J, Fall T, Ingelsson E, Thompson SG (2017) Sensitivity analyses for robust causal inference from mendelian randomization analyses with multiple genetic variants. Epidemiology 28(1):30–42. https://doi.org/10.1097/EDE.0000000000000559
Cao Y, Ai M, Liu C (2024) The impact of lipidome on breast cancer: a Mendelian randomization study. Lipids Health Dis 23(1):109. https://doi.org/10.1186/s12944-024-02103-2
Christenson SA, Smith BM, Bafadhel M, Putcha N (2022) Chronic obstructive pulmonary disease. Lancet (London, England) 399(10342):2227–2242. https://doi.org/10.1016/S0140-6736(22)00470-6
Dasgupta S, Ghosh N, Bhattacharyya P, Roy Chowdhury S, Chaudhury K (2023) Metabolomics of asthma, COPD, and asthma-COPD overlap: an overview. Crit Rev Clin Lab Sci 60(2):153–170. https://doi.org/10.1080/10408363.2022.2140329
Davey Smith G, Hemani G (2014) Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet 23(R1):R89–R98. https://doi.org/10.1093/hmg/ddu328
Davies NM, Holmes MV, Davey Smith G (2018) Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ (Clinical Research Ed) 362:k601. https://doi.org/10.1136/bmj.k601
Donnelly LE (2014) The lipidome: a new player in chronic obstructive pulmonary disease pathophysiology? Am J Respir Crit Care Med 190(2):124–125. https://doi.org/10.1164/rccm.201406-1061ED
Eggers LF, Müller J, Marella C, Scholz V, Watz H, Kugler C, Rabe KF, Goldmann T, Schwudke D (2017) Lipidomes of lung cancer and tumour-free lung tissues reveal distinct molecular signatures for cancer differentiation, age, inflammation, and pulmonary emphysema. Sci Rep 7(1):11087. https://doi.org/10.1038/s41598-017-11339-1
Gai X, Guo C, Zhang L, Zhang L, Abulikemu M, Wang J, Zhou Q, Chen Y, Sun Y, Chang C (2021) Serum glycerophospholipid profile in acute exacerbation of chronic obstructive pulmonary disease. Front Physiol 12:646010. https://doi.org/10.3389/fphys.2021.646010
GBD 2019 Chronic Respiratory Diseases Collaborators (2023) Global burden of chronic respiratory diseases and risk factors, 1990–2019: an update from the Global Burden of Disease Study 2019. EClinicalMedicine 59:101936. https://doi.org/10.1016/j.eclinm.2023.101936
Article Google Scholar
GBD Chronic Respiratory Disease Collaborators (2020) Prevalence and attributable health burden of chronic respiratory diseases, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Respir Med 8(6):585–596. https://doi.org/10.1016/S2213-2600(20)30105-3
Gould GS, Hurst JR, Trofor A, Alison JA, Fox G, Kulkarni MM, Wheelock CE, Clarke M, Kumar R (2023) Recognising the importance of chronic lung disease: a consensus statement from the Global Alliance for Chronic Diseases (Lung Diseases group). Respir Res 24(1):15. https://doi.org/10.1186/s12931-022-02297-y
Guo C, Sun L, Zhang L, Dong F, Zhang X, Yao L, Chang C (2021) Serum sphingolipid profile in asthma. J Leukoc Biol 110(1):53–59. https://doi.org/10.1002/JLB.3MA1120-719R
Hizawa N (2024) Common pathogeneses underlying asthma and chronic obstructive pulmonary disease-insights from genetic studies. Int J Chron Obstruct Pulm Dis 19:633–642. https://doi.org/10.2147/COPD.S441992
Hornburg D, Wu S, Moqri M, Zhou X, Contrepois K, Bararpour N, Traber GM, Su B, Metwally AA, Avina M, Zhou W, Ubellacker JM, Mishra T, Schüssler-Fiorenza Rose SM, Kavathas PB, Williams KJ, Snyder MP (2023) Dynamic lipidome alterations associated with human health, disease and ageing. Nat Metab 5(9):1578–1594. https://doi.org/10.1038/s42255-023-00880-1
Jiang T, Dai L, Li P, Zhao J, Wang X, An L, Liu M, Wu S, Wang Y, Peng Y, Sun D, Zheng C, Wang T, Wen X, Cheng Z (2021) Lipid metabolism and identification of biomarkers in asthma by lipidomic analysis. Biochim Biophys Acta Mol Cell Biol Lipids 1866(2):158853. https://doi.org/10.1016/j.bbalip.2020.158853
Kishimoto K, Urade R, Ogawa T, Moriyama T (2001) Nondestructive quantification of neutral lipids by thin-layer chromatography and laser-fluorescent scanning: suitable methods for “lipidome” analysis. Biochem Biophys Res Commun 281(3):657–662. https://doi.org/10.1006/bbrc.2001.4404
Kopczynski D, Hoffmann N, Peng B, Ahrends R (2020) Goslin: a grammar of succinct lipid nomenclature. Anal Chem 92(16):10957–10960. https://doi.org/10.1021/acs.analchem.0c01690
Kotlyarov S, Bulgakov A (2021) Lipid metabolism disorders in the comorbid course of nonalcoholic fatty liver disease and chronic obstructive pulmonary disease. Cells 10(11):2978. https://doi.org/10.3390/cells10112978
Lambert AA, Putcha N, Drummond MB, Boriek AM, Hanania NA, Kim V, Kinney GL, McDonald MN, Brigham EP, Wise RA, McCormack MC, Hansel NN, COPDGene Investigators (2017) Obesity is associated with increased morbidity in moderate to severe COPD. Chest 151(1):68–77. https://doi.org/10.1016/j.chest.2016.08.1432
Li L, Liu Y, Liu X, Zheng N, Gu Y, Song Y, Wang X (2022) Regulatory roles of external cholesterol in human airway epithelial mitochondrial function through STARD3 signalling. Clin Transl Med 12(6):e902. https://doi.org/10.1002/ctm2.902
Li Y, Wang W, Zhou D, Lu Q, Li L, Zhang B (2023) Mendelian randomization study shows a causal effect of asthma on chronic obstructive pulmonary disease risk. PLoS One 18(9):e0291102. https://doi.org/10.1371/journal.pone.0291102
Loureiro CC, Oliveira AS, Santos M, Rudnitskaya A, Todo-Bom A, Bousquet J, Rocha SM (2016) Urinary metabolomic profiling of asthmatics can be related to clinical characteristics. Allergy 71(9):1362–1365. https://doi.org/10.1111/all.12935
Luo J, Yang H, Song BL (2020) Mechanisms and regulation of cholesterol homeostasis. Nat Rev Mol Cell Biol 21(4):225–245. https://doi.org/10.1038/s41580-019-0190-7
Miller RL, Grayson MH, Strothman K (2021) Advances in asthma: new understandings of asthma’s natural history, risk factors, underlying mechanisms, and clinical management. J Allergy Clin Immunol 148(6):1430–1441. https://doi.org/10.1016/j.jaci.2021.10.001
Ntontsi P, Photiades A, Zervas E, Xanthou G, Samitas K (2021) Genetics and epigenetics in Asthma. Int J Mol Sci 22(5):2412. https://doi.org/10.3390/ijms22052412
Ono JG, Worgall TS, Worgall S (2015) Airway reactivity and sphingolipids-implications for childhood asthma. Mol Cell Pediatr 2(1):13. https://doi.org/10.1186/s40348-015-0025-3
Ottensmann L, Tabassum R, Ruotsalainen SE, Gerl MJ, Klose C, Widén E, Finn G, Simons K, Ripatti S, Pirinen M (2023) Genome-wide association analysis of plasma lipidome identifies 495 genetic associations. Nat Commun 14(1):6934
Pascoe CD, Jha A, Ryu MH, Ragheb M, Vaghasiya J, Basu S, Stelmack GL, Srinathan S, Kidane B, Kindrachuk J, O’Byrne PM, Gauvreau GM, Ravandi A, Carlsten C, Halayko AJ (2021) Allergen inhalation generates pro-inflammatory oxidised phosphatidylcholine associated with airway dysfunction. Eur Respir J 57(2):2000839. https://doi.org/10.1183/13993003.00839-2020
Raghu P (2020) Functional diversity in a lipidome. Proc Natl Acad Sci USA 117(21):11191–11193. https://doi.org/10.1073/pnas.2004764117
Ravi A, Goorsenberg AWM, Dijkhuis A, Dierdorp BS, Dekker T, van Weeghel M, Sabogal Piñeros YS, Shah PL, Ten Hacken NHT, Annema JT, Sterk PJ, Vaz FM, Bonta PI, Lutter R (2021) Metabolic differences between bronchial epithelium from healthy individuals and patients with asthma and the effect of bronchial thermoplasty. J Allergy Clin Immunol 148(5):1236–1248. https://doi.org/10.1016/j.jaci.2020.12.653
Reed RM, Iacono A, DeFilippis A, Eberlein M, Girgis RE, Jones S (2011) Advanced chronic obstructive pulmonary disease is associated with high levels of high-density lipoprotein cholesterol. J Heart Lung Transplant 30(6):674–678. https://doi.org/10.1016/j.healun.2010.12.010
Ried JS, Baurecht H, Stückler F, Krumsiek J, Gieger C, Heinrich J, Kabesch M, Prehn C, Peters A, Rodriguez E, Schulz H, Strauch K, Suhre K, Wang-Sattler R, Wichmann HE, Theis FJ, Illig T, Adamski J, Weidinger S (2013) Integrative genetic and metabolite profiling analysis suggests altered phosphatidylcholine metabolism in asthma. Allergy 68(5):629–636. https://doi.org/10.1111/all.12110
Sugiura H, Koarai A, Ichikawa T, Minakata Y, Matsunaga K, Hirano T, Akamatsu K, Yanagisawa S, Furusawa M, Uno Y, Yamasaki M, Satomi Y, Ichinose M (2012) Increased 25-hydroxycholesterol concentrations in the lungs of patients with chronic obstructive pulmonary disease. Respirology (Carlton, Vic) 17(3):533–540. https://doi.org/10.1111/j.1440-1843.2012.02136.x
Tibboel J, Reiss I, de Jongste JC, Post M (2014) Sphingolipids in lung growth and repair. Chest 145(1):120–128. https://doi.org/10.1378/chest.13-0967
Xuan L, Han F, Gong L, Lv Y, Wan Z, Liu H, Zhang D, Jia Y, Yang S, Ren L, Liu L (2018) Association between chronic obstructive pulmonary disease and serum lipid levels: a meta-analysis. Lipids Health Dis 17(1):263. https://doi.org/10.1186/s12944-018-0904-4
Yoder M, Zhuge Y, Yuan Y, Holian O, Kuo S, van Breemen R, Thomas LL, Lum H (2014) Bioactive lysophosphatidylcholine 16:0 and 18:0 are elevated in lungs of asthmatic subjects. Allergy Asthma Immunol Res 6(1):61–65. https://doi.org/10.4168/aair.2014.6.1.61
Yu H, Wan X, Yang M, Xie J, Xu K, Wang J, Wang G, Xu P (2023) A large-scale causal analysis of gut microbiota and delirium: a Mendelian randomization study. J Affect Disord 329:64–71. https://doi.org/10.1016/j.jad.2023.02.078
Download references
Acknowledgements
We would like to thank all the authors for their contributions to this study. We also extend our gratitude to all the researchers and volunteer participants involved in GWAS research.
This study was funded by National Natural Science Foundation of China (81973670), Key Projects of Traditional Chinese Medicine Research in Hunan Province (A2024010), Excellent Youth Project of Hunan Provincial Department of Education (23B0379), Undergraduate research and innovation fund project of Hunan University of Chinese Medicine (2023BKS153).
Author information
Qiong Wu and Jingmin Fu contributed equally to this work and share first authorship.
Authors and Affiliations
College of Humanities and Management, Hunan University of Chinese Medicine, Xueshi Road 300, Changsha, 410208, Hunan, People’s Republic of China
College of Traditional Chinese Medicine, Hunan University of Chinese Medicine, Xueshi Road 300, Changsha, 410208, Hunan, People’s Republic of China
Jingmin Fu, Jianing Shi, Zhiying Feng & Kangyu Wang
The College of Integrated Traditional Chinese and Western Medicine, Hunan University of Chinese Medicine, Xueshi Road 300, Yuelu District, Changsha, 410208, Hunan, People’s Republic of China
Cheng Zhang, Zhuolin Liu & Ling Li
You can also search for this author in PubMed Google Scholar
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Ling Li, Qiong Wu, Jingmin Fu, Cheng zhang, Zhuolin Liu, Jianing Shi, Zhiying Feng and Kangyu Wang. The first draft of the manuscript was written by Qiong Wu and Jingmin Fu, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Correspondence to Ling Li .
Ethics declarations
Conflict of interest.
The authors have no competing interests to declare that are relevant to the content of this article.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Wu, Q., Fu, J., Zhang, C. et al. Causal relationship between Lipdome and Chronic Obstructive Pulmonary Disease and Asthma: Mendelian randomization. 3 Biotech 14 , 249 (2024). https://doi.org/10.1007/s13205-024-04071-x
Download citation
Received : 04 April 2024
Accepted : 28 August 2024
Published : 25 September 2024
DOI : https://doi.org/10.1007/s13205-024-04071-x
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Mendelian randomization
- Chronic obstructive pulmonary disease
- Causal relationship
- Find a journal
- Publish with us
- Track your research

IMAGES
VIDEO
COMMENTS
Significance of the study in research refers to the potential importance, relevance, or impact of the research findings. It outlines how the research contributes to the existing body of knowledge, what gaps it fills, or what new understanding it brings to a particular field of study. In general, the significance of a study can be assessed based ...
4. Mention the Specific Persons or Institutions Who Will Benefit From Your Study. 5. Indicate How Your Study May Help Future Studies in the Field. Tips and Warnings. Significance of the Study Examples. Example 1: STEM-Related Research. Example 2: Business and Management-Related Research.
The most obvious measure of a study's long term research significance is the number of citations it receives from future publications. The thinking is that a study which receives more citations will have had more research impact, and therefore significance, than a study which received less citations.
Significance of the Study Format. When writing the "Significance of the Study" section in a research paper, follow this format to ensure clarity and impact: 1. Introduction. Contextual Background: Provide a brief background of the research topic. Research Problem: State the problem the study addresses. 2.
The significance of the study is a section in the introduction of your thesis or paper. It's purpose is to make clear why your study was needed and the specific contribution your research made to furthering academic knowledge in your field. In this guide you'll learn: what the significance of the study means, why it's important to include ...
To write a compelling significance statement, identify the research problem, explain why it is significant, provide evidence of its importance, and highlight its potential impact on future research, policy, or practice. A well-crafted significance statement should effectively communicate the value of the research to readers and help them ...
These tips will tell you the basic components expected to be seen in the significance of the study content. 1. Refer to the Problem Statement. In writing the significance of the study, always refer to the statement of the problem. This way, you can clearly define the contribution of your study. To simplify, your research should answer this ...
Answer: The significance of the study is the importance of the study for the research area and its relevance to the target group. You need to write it in the Introduction section of the paper, once you have provided the background of the study. You need to talk about why you believe the study is necessary and how it will contribute to a better ...
of the Study. The research proposal is a written docu ment which specifies what the researcher intends to study and sets forth the plan or design for answering the research ques tion(s). Frequently investigators seek funding support in order to implement the proposed research. There are a variety of funding sources that sponsor research.
Example 1: "There has been a significant increase in the incidence of diabetes in recent years. This has led to an increased demand for effective diabetes management strategies. The purpose of this study is to evaluate the effectiveness of a new diabetes management program in improving patient outcomes.".
Significance of a study is established by making a case for it, not by simply choosing hypotheses everyone already thinks are important. Although you might believe the significance of your study is obvious, readers will need to be convinced. Significance is something you develop in your evolving research paper.
The Significance of the Study describes what contribution your study will make to the broad literature or set of broad educational problems upon completion. In this activity, you will draft your Significance of the Study by determining what you hope will benefit others and/or how readers will benefit or learn from your study. As you draft your ...
What is the significance of a study and how is it stated in a research paper? The basics of writing a statement of the problem for your research proposal; 4 Step approach to writing the Introduction section of a research paper; For more information, you may search the site using the relevant keywords. Hope that helps. All the best for your study!
Step 4: Future Studies in the Field. Next, discuss how the significance of your research will benefit future studies, which is especially helpful for future researchers in your field. In the example of cyberbullying affecting student performance, your research could provide further opportunities to assess teacher perceptions of cyberbullying ...
Summary. A study's significance usually appears at the end of the Introduction and in the Conclusion to describe the importance of the research findings. A strong and clear significance statement will pique the interest of readers, as well as that of relevant stakeholders. Maximise your publication success with Charlesworth Author Services.
Signi cance of a study is established by making a case for. it, not by simply choosing hypotheses everyone already thinks are important. Although you might believe the signi cance of your study is ...
The rationale of the study is the justification for taking on a given study. It explains the reason the study was conducted or should be conducted. This means the study rationale should explain to the reader or examiner why the study is/was necessary. It is also sometimes called the "purpose" or "justification" of a study.
The significance of the study in research pertains to the potential significance, relevance, or influence of the research results. It elucidates the ways in which the research contributes to the current knowledge base, addresses existing gaps, or provides new insights within a specific field of study. Whether you are composing a research paper ...
The significance of the study must explain the importance of the work and its potential benefits. Among what makes the significance of the study plausible is that the researcher will discuss the ...
The significance of the study is a written statement (written with a non-expert in mind) that explains why your research was needed. It presents the importance of your research. It's a justification of the importance of your work and the impact it has on your research field, its contribution to new knowledge, and how others will benefit from it.
The "so what" ques-tion is one of the most basic questions, often perceived by novice researchers as the most dificult question to answer. Indeed, addressing the "so what" question contin-ues to challenge even experienced researchers. It is not always easy to articulate a convincing argument for the importance of your work.
When testing statistical significance, it's essential to: Clearly define the null and alternative hypotheses before collecting data. Choose an appropriate statistical test based on the data type and distribution. Interpret p-values in the context of the study design, sample size, and potential confounding factors.
As the largest public funder of biomedical research in the world, NIH supports a variety of programs from grants and contracts to loan repayment. Learn about assistance programs, how to identify a potential funding organization, and past NIH funding. ... Biosketch Format Pages, Instructions, and Samples. Scope Note. A biographical sketch (also ...
Answer: In simple terms, the significance of the study is basically the importance of your research. The significance of a study must be stated in the Introduction section of your research paper. While stating the significance, you must highlight how your research will be beneficial to the development of science and the society in general.
The interpretation of large datasets, such as The Cancer Genome Atlas (TCGA), for scientific and research purposes, remains challenging despite their public availability. In this study, we focused on identifying gene expression profiles most relevant to patient prognosis and aimed to develop a method and database to address this issue. To achieve this, we introduced Luo's Optimization ...
The significance of the study, quite simply, is the importance of the study to the field - what new insights/information it will yield, how it will benefit the target population, very simply, why it needs to be conducted. For instance, given the current situation (and without knowing your subject area), you may wish to conduct research on ...
Background Oligotrophy and hypereutrophy represent the two extremes of lake trophic states, and understanding the distribution of bacterial communities across these contrasting conditions is crucial for advancing aquatic microbial research. Despite the significance of these extreme trophic states, bacterial community characteristics and co-occurrence patterns in such environments have been ...
This study validates the relationship between lipdome and these diseases through large-scale GWAS data. MR, as an important method in genetic research, has great epidemiological significance. This study also further complements the previous lipidomics views on lung-related diseases and provides some ideas for clinical diagnosis and treatment.