What Is Data Transcription Really? Understand Qualitative Data Transcription to Get Ahead
Rev › Blog › Resources › Other Resources › Definitions & Glossaries › What Is Data Transcription Really? Understand Qualitative Data Transcription to Get Ahead
What do you think of when you hear the word “data”? Most people might immediately think of measurements, statistics, and rows of numbers. You might think of computers or even robots collecting and calculating information and spitting out answers based on cold, unchanging facts.
Actually, that is just one of a few different types of data. Quantitative data is the type of data that can be expressed in numbers and precisely measured, like temperature or population size. But that’s not the only type of data that scientists and other professionals use in research.
A second type of data is known as qualitative data . Qualitative data is subjective, open-ended, and less precise in how it is measured or analyzed. This type of data is expressed in words, rather than numbers. Qualitative data analysis describes qualities or characteristics, and may be expressed through opinions or preferences.
For example, companies may use qualitative data to understand the decisions or preferences of different consumers or consumer groups. Medical scientists may look at qualitative data to understand how or why patients make certain health decisions.

Why use transcription for qualitative data?
Qualitative research is more about exploring an idea or a topic instead of finding specific, concrete, objective answers. Since qualitative research focuses on individuals, groups, and cultures, its data can’t be measured with tools like thermometers and scales. Instead, qualitative data is measured with questionnaires, observations, or interviews. All this can make qualitative data more difficult to record and copy compared with quantitative data.
Qualitative researchers are focused on understanding a person’s opinion or why people behave in certain ways. This means that researchers may conduct and record focus groups, group discussions, individual interviews, or observations of a person or group of people. They may capture and preserve the resulting data with video or audio recordings.
These interviews and other events create important data. However, that data is usually unstructured and needs to be sorted through and organized before researchers can make sense of it.
This is where qualitative data transcription is incredibly important. Transcription creates a text-based version of any original audio or video recording. Qualitative data transcription provides a good first step in arranging your data systematically and analyzing it.
Transcription is vital for qualitative research because it:
- Puts qualitative data and information into a text-based format
- Makes data easier to analyze and share
- Allows researchers to become more immersed into the data they collect
- Helps researchers create a narrative with their data
- Makes patterns easier to find
- Helps preserve the accuracy and integrity of the data
- Lets researchers focus on their observations instead of worrying about note-taking
Once data is transcribed in a text format, it can be put into a spreadsheet or similar type of document, or entered into a qualitative data analysis tool. After data transcription, a qualitative researcher can read through and annotate the transcriptions, then conceptualize and organize the data to conduct inductive or deductive analysis . From there, it is a lot easier to make connections between different observations or findings, and then write them up in the form of a study, report, or article.
Using Rev Transcription for Qualitative Data Analysis and Research
If you’re a researcher, you know how time-consuming each step of the process can be. Qualitative analysis and research involves a lot of hard work that generates a lot of data to be recorded, organized, and analyzed.
If you’re looking for any way to shorten or speed up the process without sacrificing the integrity of your research, hiring someone else to transcribe your recordings is vital. Using a professional outside transcription service can help you streamline your process, save time, and ensure that you preserve the quality and usability of the original recordings you capture.
Rev’s transcription services not only help you capture more value from your recorded audio and video, but also offer a smooth, easy, online experience. Our human, native English-speaking transcriptionists can capture the details of your interviews, conversations, and observations, so you can put more attention where it’s needed.
It’s easy to get started with Rev transcription:
- Securely upload files from your computer or paste a URL from the web . Add any notes, like a glossary or speaker names, that you think may help the transcriptionist.
- Choose your payment method. Rev.com transcription starts at $1 per audio minute, although you can add features like verbatim transcription or rush delivery.
- Let our 50,000+ professionals get started transcribing your files. We promise 99% accuracy or better, 100% guaranteed. You’ll get your transcript back as a document you can view and edit with our tools, then share with anyone.
No matter the day of the week, we provide quick transcription turnarounds. Standard delivery is at least 12 hrs for most files under 30 minutes, although the exact time will depend on the length of the recording and the quality of the audio.
Here at Rev, we require our transcriptionists to meet rigorous quality standards before working on customer files. We also have a team of reviewers that works with the transcriptionists to ensure that your files are ready for delivery.
Concerned about your data confidentiality? You don’t have to be. Rev also has a strict customer confidentiality policy that keeps files private and protected from unauthorized access. All Rev professionals have signed NDAs and strict confidentiality agreements, and can only complete work on our secure platform. Have your own NDA you want us to sign? We can do that too; just contact us .
Start streamlining your qualitative research process and capture more accurate data faster by trying Rev today .
More Caption & Subtitle Articles
Everybody’s Favorite Speech-to-Text Blog
We combine AI and a huge community of freelancers to make speech-to-text greatness every day. Wanna hear more about it?
University Library, University of Illinois at Urbana-Champaign

Qualitative Data Analysis: Transcription
- Atlas.ti web
- R for text analysis
- Microsoft Excel & spreadsheets
- Other options
- Planning Qual Data Analysis
- Free Tools for QDA
- QDA with NVivo
- QDA with Atlas.ti
- QDA with MAXQDA
- PKM for QDA
- QDA with Quirkos
- Working Collaboratively
- Qualitative Methods Texts
- Transcription
- Data organization
- Example Publications
- Find Methods Examples
Transcription as an Act of Analysis
While transcription is often treated as part of the data collection process, it is also an act of analysis (Woods, 2020). When you manually transcribe an interview, for example, you make choices about how to turn the recording of the interview into text, and these decisions shape the analysis you conduct.
For example, if you host a focus group, a transcription that just includes the words spoken by the participants loses data about the interaction between them. You may decide to ensure that your transcription includes details on interactions (which would take more time or resources) or decide that interaction information is not relevant to your analysis. This decision is influenced by your methodology and research goals, and should be recognized as a part of your analysis process.
Planning and communicating the transcription process is further complicated when the researcher works in a research team, asks participants to discuss sensitive topics, occurs in a cross-cultural environment, or when the transcript must be translated into another language (Clark et al, 2017). Published research reports rarely include significant detail about the transcription process, but if you find yourself in one of these situations, it may be worth seeking works in your discipline that address best practices for transcription, data management, participant relationships, and translation, such as Clark et al's (2017) work on developing a transcription and translation protocol for sensitive and cross-cultural team research.
Transcription Tools
- Atlas.ti (Mac)
- Atlas.ti (Windows)
- NVivo (Windows)
- NVivo (Mac)
- Kaltura/Mediaspace
- Free transcription tools
- Paid transcription services
- Importing automatic transcripts into Atlas.ti (Mac) You can import transcripts and media files from Zoom, Teams, and other video meeting platforms. Atlas.ti links the video and automatic transcript, which allows you to watch the video and edit the transcript right in Atlas.
- Creating transcripts in Atlas.ti You can import media files to Atlas.ti and then create your own transcript within the program. This process will create a transcript that is synced with the media file.
- Link a transcript to media in Atlas.ti You can import existing media and transcripts to Atlas.ti in order to link them together and enable synchronous viewing of the media with links to the transcript.
- Importing automatic transcripts into Atlas.ti (Windows) You can import transcripts and media files from Zoom, Teams, and other video meeting platforms. Atlas.ti links the video and automatic transcript, which allows you to watch the video and edit the transcript right in Atlas.
- Create a transcript in Atlas.ti You can import media files to Atlas.ti and create your own transcript within the program. The transcript will be linked to the media for synchronous scrolling.
- Link a transcript to a media file If you transcript text already, you can upload a media file to Atlas.ti and link the text. This will allow you to use synchronized scrolling, which shows you the video and transcript at the same time.
- Create transcripts in NVivo You can create new transcriptions of media in your NVivo project using the edit mode.
- Import and link transcripts in NVivo Existing transcripts can be imported to NVivo and link the transcript with a media file.
The MAXQDA is the same across Mac and Windows devices.
- Manual Transcription You can upload media files to MAXQDA and then create new transcripts using the Multimedia Browser.
- Link transcripts to a media file by creating timestamps If you already have transcript text, you can use the edit mode in MAXQDA to create timestamps and sync the transcript to the media file.
- Automatic transcription New to MAXQDA 24, you can now automatically transcribe your media.
- Downloading captions from Kaltura Video files you upload to Kaltura (including recorded Zoom meetings) are automatically captioned, though you'll need to edit the captions and publish them before they appear on your video. Once the file is created, you can download it from Kaltura to upload to other programs. See this page on captions in Kaltura for more information.
- Find and replace text in Word When you download captions from Zoom or Kaltura, it will come with timestamps. You can use the find and replace feature in Word to clear the timestamps for easier editing.
- OTranscribe OTranscribe is a free, open-source and web browser based tool for transcribing audio and video. You can upload media and use the tool to create citations. See the help pages for information.
- Google Docs Voice Typing You can use the voice typing feature to create rough transcriptions of audio as you collect data or by re-playing a recording into the microphone.
- Microsoft Word Dictate Typing Web and desktop versions of Microsoft Word include a dictation tool that will create a rough transcription while you collect data or when you play a recording near your device's microphone.
There are companies that will create transcripts from media files on your behalf, usually for a by-minute fee.
If you decide to use one of these options, you should ensure that the security of data shared with these services is in compliance with your IRB protocol and consent obtained from any participants.
Do you have experience with any paid transcription services that you think would be worth adding to this list? Please share your experience with me .
- NVivo Transcription NVivo offers a paid transcription service, which can be purchased as a paid subscription or a pay-as-you-go service. Transcription is available in 42 languages including English, Spanish, Japanese, Hindi, Arabic and Korean.
- Rev Ref offers both automatic, rough transcription as well as more accurate transcription conducted by workers. Rev supports 36 languages including English, Spanish, Arabic, Mandarin, Japanese, Korean, and Hindi.
- Trint Trint is a paid transcription and analysis tool, with transcription available for 30+ languages , including English, Spanish, Chinese Mandarin, Korean, Hindi, and Korean. Trint also offers translation of text.
Cited on this page
Clark, L., Birkhead, A. S., Fernandez, C., & Egger, M. J. (2017). A transcription and translation protocol for sensitive cross-cultural team research . Qualitative Health Research , 27 (12), 1751–1764. https://doi.org/10.1177/1049732317726761
Woods, D. Presentation in: Christina Silver, Phd. (2020, December 4). CAQDAS webinar 005 Transcription as an analytic act. [Video]. https://www.youtube.com/watch?v=7X-s1r4l0QQ.
- << Previous: Qualitative Data Analysis Strategies
- Next: Data organization >>
- Last Updated: Jun 6, 2024 9:59 AM
- URL: https://guides.library.illinois.edu/qualitative

The Ultimate Guide to Qualitative Research - Part 2: Handling Qualitative Data

- Handling qualitative data
- Introduction
Introduction to transcripts in qualitative research
Understanding the transcription process, practical insights: transcription in action, using transcription services, challenges in transcription.
- Field notes
- Survey data and responses
- Visual and audio data
- Data organization
- Data coding
- Coding frame
- Auto and smart coding
- Organizing codes
- Qualitative data analysis
- Content analysis
- Thematic analysis
- Thematic analysis vs. content analysis
- Narrative research
- Phenomenological research
- Discourse analysis
- Grounded theory
- Deductive reasoning
- Inductive reasoning
- Inductive vs. deductive reasoning
- Qualitative data interpretation
- Qualitative analysis software
Research transcripts
Conducting qualitative interviews or focus groups is only the first part of data collection in a qualitative research project. For most qualitative data analysis , you need to turn those audio or video files into written transcripts. While this may seem self-evident to many researchers, much discussion has taken place about transcripts, best research practices for generating them, the debate between transcription services and human transcription, and so much more.
Qualitative data transcription holds a key role in research , acting as the building blocks from which findings are derived and conclusions are drawn. They are the textual representation of verbal data gathered through interviews , focus groups , and observational studies . Given their significance, it's essential to grasp why they are fundamental to qualitative research.
What is the importance of transcripts in research?
The importance of transcripts in research lies in their ability to convert spoken language into written form, making data analysis significantly more manageable. Transcripts act as the raw material for your analysis , creating a tangible record of the conversations and discussions that form the basis of your research. They provide a precise, detailed account of the verbal data collected, enabling researchers to review the information repeatedly and uncover layers of meaning that might be overlooked when listening to the recording .
Transcripts help researchers systematically organize and manage the data, especially when dealing with large volumes of information. They make it easier to search for specific themes, patterns, or keywords, thereby speeding up the data analysis process. Furthermore, transcripts facilitate the sharing of data among researchers, allowing for collaborative analysis and review. They also ensure the transparency of your research by providing a permanent record that can be scrutinized by other researchers, reviewers, or auditors.
How is transcribing used in qualitative research?
A transcript is used as a way to record and represent the rich, detailed, and complex data collected during qualitative studies such as interviews, focus groups, or observations. Without transcriptions, it would be challenging for researchers to dissect, understand, and interpret the in-depth experiences, perceptions, and opinions shared by the participants. Most research involving audio recordings of interviews requires recordings to undergo the transcription process in order for qualitative data analysis to proceed.
Transcribing, in qualitative research, doesn't merely involve verbatim transcription (the word-for-word rendering of verbal data into text). It can also encompass the translation of non-verbal cues such as laughter, pauses, or emotional expressions that can provide valuable context and insights into the participants' experiences and perspectives. By capturing these details, transcripts can help portray a fuller, more authentic picture of the data, enabling a more comprehensive and nuanced analysis.
In qualitative research, transcriptions are also used for data coding , a process where researchers label or categorize parts of the data based on their content, themes, or patterns. This step is critical for identifying trends and making sense of the data, and having a written transcript makes the coding process significantly more efficient and precise.
How are transcripts used in quantitative research?
Interview transcripts also have an important role in quantitative research , specifically in methods like content analysis and conversation analysis . Content analysis involves the systematic coding and quantifying of data within transcripts, such as the frequency of specific words or themes. This allows researchers to discern patterns and trends and gain insights into the prevalence of certain concepts or attitudes. For example, this could involve quantifying the occurrence of health-related discussions within interviews with healthcare providers.

On the other hand, conversation analysis , while often qualitative, can include quantifiable aspects. Transcripts record details of conversation structure and patterns, such as timing and sequence of speech. Quantitative measures like the count of certain conversational elements or the duration of pauses can be used to understand communication dynamics.
In essence, transcripts are not solely a tool for qualitative research methods but also provide a source of quantitative data and a foundation for quantitative analysis methods. They allow for a detailed, tangible record of spoken data, crucial for both qualitative understanding and quantitative measures, showcasing their versatility in the research field.
The transcription process is a critical stage in qualitative research . It refers to the conversion of recorded or observed speech into written text, turning the fluid and dynamic nature of spoken communication into a tangible and analyzable form . In this section, we will delve deeper into the process of transcription and how it is approached in qualitative research.
How do you create a research transcript?
Writing a research transcript starts with the raw data , usually an audio or video recording from interviews , focus groups , or observations . The first step is to carefully listen to the recording and begin writing down what is being said. This should be done with utmost accuracy, capturing not only the spoken words but also any significant pauses, laughter, or emotional expressions.
A crucial aspect of writing a transcript is deciding how detailed it should be. This varies depending on the research objectives and the nature of the data. For some research, a verbatim transcription, which includes every utterance, filler words, and non-verbal cues, is necessary. For other studies, a clean verbatim transcript, which omits irrelevant details like repeated words or stutters, is sufficient. After the initial transcription, the transcript should be reviewed and cross-checked with the recording for accuracy. During this revision process, the researcher may also add time stamps, annotations, or comments to enrich the transcript further.
Other details in transcripts
Depending on your research inquiry, you may consider more nuanced approaches to generating transcripts when you require the analysis of complex and multifaceted data. Apart from accurately rendering the spoken words into text, a qualitative research transcript can also capture the context, meaning, and nuances inherent in the spoken interaction.
This could involve noting the tone of voice, pauses, emotional expressions, body language, and interactions among participants. These non-verbal cues can provide rich insights into the participants' attitudes, emotions, and social dynamics, thus giving the researcher a deeper understanding of the data.
One unique aspect of transcribing qualitative data is the reflection and interpretative process embedded in it. Researchers often gain a deeper understanding of the data during transcription, as it forces them to engage closely with the data and notice details that might have been missed during the initial data collection.
How is data transcription done?
Data transcription can be done manually or with the assistance of transcription software. Manual transcription involves the researcher or a transcriptionist listening to the recording and typing out the conversation. This method is time-consuming but can lead to a higher level of accuracy and deeper immersion in the data.
Automated transcription software, on the other hand, uses automatic speech recognition (ASR) technology to transcribe audio recordings into text. While this method is faster and can handle large volumes of data, it may not be as accurate, especially when dealing with poor audio quality, heavy accents, or technical jargon.
Regardless of the method chosen, the transcribed data should be reviewed and edited for accuracy. This might involve repeated listening to the audio, making corrections, and refining the transcript until it accurately represents the original data.
In summary, the transcription process is a meticulous task that requires careful listening, accurate writing, and thoughtful interpretation. It is an essential step in transforming the raw data into a form suitable for in-depth analysis, thus laying the foundation for your qualitative research findings. By understanding how to write a research transcript, specifically a qualitative research transcript, and knowing how data transcription is done, you'll be well-equipped to handle this critical phase of your qualitative research process.
Types of data transcription in qualitative research
As qualitative data can be diverse and complex, it’s important to understand that not all transcripts are the same. Depending on the research objectives, data characteristics, and the resources available, researchers might opt for different types of transcriptions. Let's delve deeper into these different types and their applicability in qualitative research.
What are the different types of data transcription?
There are generally three main types of data transcription:
1. Verbatim transcription: This is the most detailed form of transcription. It involves transcribing every single word, including filler words (like "um," "uh," and "you know"), false starts, repetitions, and even non-verbal cues such as laughter, pauses, or sighs. Verbatim transcription is often used in research where the manner of speaking or the emotional context is as important as the content itself.
2. Clean verbatim transcription: This type of transcription also captures every word spoken but omits filler words, stutters, and false starts, resulting in a cleaner, more readable transcript. Clean verbatim transcription is usually preferred when the focus is on the content of the speech rather than the style or manner of speaking.
3. Intelligent transcription (or edited transcription): This form of transcription goes a step further in simplifying and clarifying the text. It not only removes filler words and repetitions but also corrects grammatical errors and may even rephrase sentences for clarity. Intelligent transcription is typically used for creating transcripts intended for publication or for audiences who are not directly involved in the research.
What are the different types of transcription in qualitative research?
In qualitative research, the type of transcription used often depends on the nature of the study and the level of detail required in the analysis.
For studies aiming to explore the content of the conversations, clean verbatim or intelligent transcriptions might be sufficient. These types provide a clear and concise account of the spoken data, allowing researchers to easily identify themes and patterns in the content.
However, for studies interested in the nuances of communication, such as sociolinguistic studies or discourse analysis, a verbatim transcription might be more appropriate. This type captures the exact words, speech patterns, and non-verbal cues, thus providing a richer and more authentic representation of the spoken interaction.

Choosing the right type of transcription for your qualitative research is crucial, as it can significantly impact the depth and quality of your data analysis . By understanding the different types of data transcription and their uses in qualitative research, you will be better positioned to make an informed decision that aligns with your research goals.
ATLAS.ti makes conducting qualitative research easy
Turn your research data into key insights starting with a free trial of ATLAS.ti.
Transcription is more than a technical process; it's a fundamental part of the journey from data collection to analysis in qualitative research . Understanding transcription in action means knowing how to do it, what to include, and how to record it for optimal use in your study.
What are examples of transcription?
Transcription can take various forms based on the nature of your research. For instance, a sociolinguistic study might require a detailed verbatim transcript, including non-verbal cues and speech anomalies.
Here's an example:
Interviewer: So, how are you feeling about the project? (in a concerned tone) Participant: Umm... Well, (laughs nervously) it's been a bit... um, overwhelming?
On the other hand, an interview transcript for a market research study might be a clean verbatim transcript, focusing on the content. Here's how it could look:
Interviewer: What do you like about our product? Participant: I really enjoy its user-friendly interface and the customer service is exceptional

How do you transcribe a research interview?
Transcribing a research interview involves several steps. First, ensure you have a good-quality audio or video recording of the interview . Listen to the recording carefully, typing out the conversation verbatim. You can also slow down the speed of the recording, and shortcut keys to rewind the recording a few seconds can be a great help. It's essential to maintain accuracy and include key details that might influence the interpretation of the data , such as significant pauses or emotional inflections.
Depending on your research aims, you may choose to transcribe in verbatim, clean verbatim, or intelligent transcription style. Once the initial transcription is complete, review and cross-check it against the recording for accuracy. Finally, anonymize the data if necessary to ensure participant confidentiality .
What should be included in an interview transcript?
An interview transcript should include everything that is said in the interview, but the level of detail can vary. Here are some elements that are typically included:
1. Identifiers: These help distinguish between different speakers. In the case of an interview, this would usually be the interviewer and the interviewee(s). 2. Verbal responses: All responses to the interview questions should be included in the transcript. 3. Non-verbal cues: Depending on the research objectives, non-verbal cues such as laughter, sighs, or pauses can provide additional context and should be included. 4. Time stamps: These help locate specific parts of the audio recording and can be very helpful during analysis. 5. Annotations: These might include comments or notes made by the transcriber about the context, the tone of voice, or background noises.

How do I record an interview transcript?
Recording an interview transcript starts with creating an audio or video recording of the interview. After the interview, use either manual transcription or automatic transcription software to convert the audio into written text. Make sure to include identifiers for each speaker, their verbal responses, and any relevant non-verbal cues. Review and revise the transcript for accuracy, adding time stamps or annotations as needed.

In summary, transcribing interviews is a meticulous task that requires careful attention to detail and accuracy. By understanding what to include in a transcript and how to record it, you'll be well-equipped to capture the richness and depth of your interview data, laying the groundwork for a robust analysis.
Benefits of transcription in qualitative research
In qualitative research , transcription represents more than a technical or administrative task. It’s the transformative process that turns spoken communication into a tangible, accessible text form that can be critically examined, dissected, and evaluated. This process forms the underpinning of the entire data analysis journey, creating the foundation upon which interpretations are built and conclusions are drawn.
Looking deeper into the benefits of transcription in qualitative research
Unearthing the multiple layers of transcription’s benefits in qualitative research reveals how it contributes to the efficacy and integrity of a study.
1. Facilitating data accessibility: One of the fundamental benefits of transcription is that it brings to life the spoken word, facilitating accessibility. It translates data into a format that is readable, searchable, and conducive to rigorous analysis. Transcripts can be reviewed multiple times, allowing researchers to revisit the data continually. They can be easily shared among team members or other researchers, enhancing the communicability of the study. Transcription also bridges barriers for those who are hearing-impaired or for whom the original language of the conversation might be a hurdle.
2. Enabling comprehensive analysis: Transcripts are the bedrock upon which qualitative analysis is built. They provide the raw material for various methods of qualitative data examination, whether it's the deep dive of a thematic analysis , the linguistic focus of discourse analysis , or the systematic categorization of content analysis . These written records allow researchers to delve into the data, identify recurring patterns, extract significant themes, and uncover insights that might be less discernible or entirely lost in the original audio or video format.
3. Promoting reflection and interpretation: Transcription is far from being a mechanical, dispassionate process. It necessitates active and continual engagement with the data , leading to a process of reflection and interpretation that forms the basis of qualitative analysis. During the act of transcribing, researchers can glean new insights, recognize overlooked details, and begin to make initial interpretations. It's often during this process that the data begin to speak, allowing researchers to discern their meaning and value.
4. Providing evidence and establishing an audit trail: Transcripts constitute a concrete, verifiable record of the data collected, the words expressed by the participants, their sentiments, and their experiences. This record acts as a form of evidence to substantiate the research findings, ensuring their credibility. Furthermore, they provide an audit trail, contributing to the transparency, accountability, and, thus, the overall trustworthiness of the study.
Justifying the use of transcription for qualitative data
The crucial role of transcription in qualitative research is underscored by its ability to capture the richness and multifaceted nature of spoken data and convert it into a format ripe for in-depth analysis. It provides a lens through which subtle nuances of communication - the ebb and flow of conversation, shifts in tone, or emotional expressions - can be understood. This is invaluable in qualitative research, where the aim is to capture and understand the depth and complexity of human experiences.
Transcripts also serve as a durable, enduring record of the data, preserving the words and voices of the participants. They ensure that the insights, stories, and experiences shared by participants are not transient but can be revisited, reviewed, and reinterpreted in future research.
Transcription services have revolutionized the way researchers process their data, offering a range of possibilities from manual transcription to advanced AI-driven software. These services often come with their own benefits and drawbacks, and understanding these is key to making an informed decision for your qualitative research project. This section will delve into the world of transcription services, helping you to explore your options and make the best choice for your research needs.
Types of transcription services
Broadly, transcription services fall into two main categories: human services and automated services.
Human transcription services employ professional transcribers to convert your audio or video files into text. These services often offer high-quality, accurate transcripts, as they benefit from the nuanced understanding and context interpretation abilities of a human transcriber.

Automated transcription services, on the other hand, use speech recognition software to transcribe audio or video files. They are typically faster and less expensive than human transcription services, but their accuracy can vary depending on the quality of the audio and the complexity of the language used.
Advantages and disadvantages of outside services
Choosing between human and automated transcription services often depends on your project's specific needs. Let's delve into some advantages and disadvantages of each.
Advantages of human services
1. Accuracy: Human transcribers can understand context, decipher accents, and make out words in poor-quality audio better than any software, ensuring high-quality transcripts.
2. Personalized service: They offer personalized service with attention to detail, including specific formatting requests or specialized transcription styles.
Disadvantages of human services
1. Time-consuming: Human transcription is slower than automated transcription, which can be an issue for projects with tight timelines.
2. Cost: Human transcription services can be expensive, especially for large volumes of data. Advantages of automated services
1. Speed: Automated services can transcribe audio or video files much faster than human transcribers.
2. Cost: They are usually more affordable than human transcription services, making them a good option for budget-conscious projects.
Disadvantages of automated services
1. Accuracy: While speech recognition technology has improved significantly, it still struggles with accents, poor audio quality, and complex terminology, which may lead to less accurate transcripts.
2. Lack of context: Automated services may not capture nuances in language or understand context the way a human transcriber can.
Tips for choosing the right service
Selecting the right transcription service should be based on the specific needs and constraints of your project. Here are a few tips to guide your choice:
1. Assess your needs: Consider the complexity of your data, the quality of your recordings, your budget, and your timeline.
2. Test the service: If possible, use a short sample of your data to test the service. This can give you a sense of the quality of the transcription and whether it meets your needs.
3. Read reviews: Check out reviews and ratings from other users to gauge the reliability and performance of the service.
These outside services can be a valuable resource in qualitative research, saving you time and effort. By understanding the benefits and drawbacks of human and automated services and evaluating your specific research needs, you can make an informed choice that best supports your research goals.
The transcription process, while invaluable to qualitative research , does not come without its fair share of challenges. The transformation of oral data into written format can be a complicated endeavor, particularly in cases where the audio quality is poor, speakers have heavy accents, or the conversation is filled with technical or specific jargon. Despite these hurdles, there are various strategies that can help you navigate these issues and ensure high-quality, accurate transcripts.
Audio quality
One of the most common challenges in transcription is dealing with poor audio quality. Background noise, low speaking volumes, or unclear pronunciations can make it difficult to distinguish what is being said. It's a good idea to invest in high-quality recording equipment and choose a quiet, controlled environment for your interviews or focus groups. Ensure that all participants speak clearly and loudly enough to be heard. If your data is already collected and the audio quality is poor, consider using noise-canceling software or hiring a professional transcription service that specializes in handling poor-quality audio.
Accents and dialects
Dealing with heavy accents or unfamiliar dialects can be challenging, particularly for automated transcription services that may not be programmed to handle a wide range of accents or dialects. Human transcribers can spend time familiarizing themselves with the accent or dialect to aid their comprehension. In some cases, it may be beneficial to engage a local transcriber who is familiar with the accent or dialect. For automated services, choosing a service that offers multilingual support or can handle a variety of accents can improve the accuracy of your transcripts.
Technical jargon and specific language
Transcribing conversations that include technical jargon, specific terminology, or industry-specific language can be a challenge, especially if the transcriber is not familiar with the terminology. If you are outsourcing your transcription to a human service, providing a glossary of terms to your transcriber can be very helpful. This can include definitions of technical terms, acronyms, or any specific language used in your study. If using an automated service, choose one that has capabilities to learn and adapt to specific terminology.
Time and resources
Transcription can be a time-consuming and resource-intensive process, especially for large volumes of data. Consider using transcription software or outsourcing to a transcription service to save time. If you’re transcribing manually, developing a systematic approach can increase efficiency. This can include using transcription software to speed up or slow down the audio, utilizing keyboard shortcuts, or creating a consistent formatting system.
Choose ATLAS.ti for your interview research
Analyze transcripts for interviews and focus groups with ATLAS.ti. Download a free trial today.
Qualitative Data Analysis
- Choosing QDA software
- Free QDA tools
- Transcription tools
- Social media research
- Mixed and multi-method research
- Network Diagrams
- Publishing qualitative data
- Student specialists
General Information
For assistance, please submit a request . You can also reach us via the chat below, email [email protected] , or join Discord server .
If you've met with us before, tell us how we're doing .
Service Desk and Chat
Bobst Library , 5th floor
Staffed Hours: Summer 2024
Mondays: 12pm - 5pm Tuesdays: 12pm - 5pm Wednesdays: 12pm - 5pm Thursdays: 12pm - 5pm Fridays: 12pm - 5pm
Data Services closes for winter break at the end of the day on Friday, Dec. 22, 2023. We will reopen on Wednesday, Jan. 3, 2024.
NYU IRB Protocol Regarding Transcription Software
Suggestions for best practice for 3rd Party Transcription:
- Researcher that have an individual other than themselves transcribe audio/video media, should let the IRB know who they are and have the transcriber sign a confidentiality agreement (Transcription Agreement Form for Individual)
- Researcher that have an on-line transcription software companies transcribe audio/video media, should follow the attached protocol established by PRIM&R (Public Responsibility in Medicine and Research,) the national organization for IRB's. The PRIM&R IRB forum offers suggestions regarding data security with a third party vendor. (see attached)
Suggestions for best practice for Data Storage:
- NYU IRB also suggests that once the transcription is complete, the dictation/interviews should be destroyed/wiped clean/deleted after they are reviewed by the researcher.
- It is not advised to use NYU Stream to store your material.
- PRIM&R (Public Responsibility in Medicine and Research) suggestions re data security with a third party vendor
- Transcription Agreement for Individual
NYU Accommodation Software
NYU Digital Accessibility Program- Read&Write
Text to Speech is a downloadable software that reads aloud what’s on the screen, which can be used to proofread transcriptions done manually or through a third-party transcription service.
NYU Adaptive Computing - Speech to Text Software Accommodations
- Kurzweil 1000 - reads books and other print materials aloud
- CCTV - closed circuit enlargement of print materials
- JAWS - allows users to listen to the contents of the computer’s screen
- ZoomText - magnification and screen reading
- Dragon Naturally Speaking - allows users to dictate, rather than type
The adaptive computing rooms at NYU make it easier for researchers with special accommodations to choose from a variety of mediums to ensure a smooth transcription process. *If you do not have special accommodations through the NYU Moses Center, please go to the above website and locate the “Inquire about these rooms” button to discuss your research needs with the computer center. For more information about the library services, please visit the website above or contact Lauren Kehoe of Lib accommodations at [email protected] or (212) 998 2509.
Suggested Transcription Resources
- Transcription Overview
- Microsoft Office 365
- Manual Transcription in MAXQDA
- Manual Transcription in ATLAS.ti
For qualitative researchers, transcribing is an integral element to the research process. There are a variety of ways that researchers can approach transcription and the preparation of qualitative data for analysis. While many researchers transcribe interviews manually there are also a variety of resources that can be used in conjunction to speed up the transcription process while retaining the accuracy of manual transcription. The following resources will highlight some considerations for you as you begin the process of preparing and cleaning your qualitative data.
- NYU Office 365 Access NYU Provides access to Microsoft Office 365 free of charge for students, staff, and faculty. This link takes you to the sign up page and provides more information on access to Office 365.
- Transcribe in Office 365 Microsoft Office 365 now includes free features for Speech-to-Text and Auto-transcription of uploaded files. This link will take you to the Microsoft support pages where you can find step-by-step instructions for using these features.

How To Order a Transcript on NYU Stream
- To order closed captions/ a transcript of your recording, simply add new media by uploading your project >> "Add New".
- Once it is uploaded, click on the title of the media and it will take you to an editing page.
- From here. You will see a drop down menu choice under the title "Actions" in which you may choose the option to "+Order Captions ". It will take approximately 3 times the length of you media to transcribe (ie 5 min film will take 15 mins) you will note that the status will change from pending to in progress to complete.
- Once your caption request is processed, click the "Actions" button, dropdown and select "Edit." Select the “Captions” option in the toolbar>Edit Captions. Edit captions as necessary while streaming the video on the righthand side of your screen.
- To download captions, return to the toolbar>Captions>select the rightmost icon under Actions.
- NYU Stream Tutorials
- MAXQDA Transcription Mode MAXQDA provides a built-in editor for manual transcription. This link will take you to the step-by-step instructions on how to use this feature.
- Transcription in ATLAS.ti 9 Learn more about preparing transcripts directly in ATLAS.ti 9 for ease of preparation and analysis.
ATLAS.ti 9 has incorporated new features that allow users to edit transcripts directly in the user interface.
Bibliographic Resources
Branley, D. (2004). 'Making and Managing Audio Recordings' in Clive Seale (ed) Researching Society and Culture (2nd edition), London: Sage, pp 207-223
Lapadat, J., & Lindsay, A. (1999). 'Transcription in Research and Practice : from standardisation of technique to interpretive positionings' Qualitative Inquiry 5(1) 64-86
Lewins, A. (1998). ' Transcribing Evaluation data ' in Harvey J. (Ed) Evaluation Cookbook Online , Learning Technology Dissemination Initiative, Herriot Watt Edinburgh.
Stockdale, A. (2003). An Approach to Recording, Transcribing, and Preparing Audio Data for Qualitative Data Analysis Education Development Center, Inc.
- << Previous: Special topics
- Next: Social media research >>
- Last Updated: May 23, 2024 2:40 PM
- URL: https://guides.nyu.edu/QDA
Transcription and Qualitative Methods: Implications for Third Sector Research
- Research Papers
- Published: 10 September 2021
- Volume 34 , pages 140–153, ( 2023 )
Cite this article

- Caitlin McMullin ORCID: orcid.org/0000-0002-7029-9998 1
74k Accesses
54 Citations
23 Altmetric
Explore all metrics
While there is a vast literature that considers the collection and analysis of qualitative data, there has been limited attention to audio transcription as part of this process. In this paper, I address this gap by discussing the main considerations, challenges and implications of audio transcription for qualitative research on the third sector. I present a framework for conducting audio transcription for researchers and transcribers, as well as recommendations for writing up transcription in qualitative research articles.
Similar content being viewed by others

Transcription and Data Management

Transcribe or Not Transcribe?
Longform recordings of everyday life: Ethics for best practices
Avoid common mistakes on your manuscript.
Introduction
The field of third sector studies is inherently interdisciplinary, with studies from political science, management, sociology and social work, among others. Within the field of research, a large percentage (between 40–80%) of studies employ qualitative methods such as interviews, focus groups and ethnographic observations (von Schnurbein et al., 2018 ). In order to ensure rigor, qualitative researchers devote considerable time to developing interview guides, consent forms and coding frameworks. While there is a vast literature that considers the collection and the analysis of qualitative data, there has been comparatively limited attention paid to audio transcription, which is the conversion of recorded audio material into a written form that can be analyzed. Despite advances made in qualitative methodologies and increasing attention to positionality, subjectivity and reliability in qualitative data analysis, the transcription of interviews and focus groups is often presented uncritically as a direct conversion of recorded audio to text. As technology to facilitate transcription improves, many researchers have shifted to using voice-to-text software and companies that employ AI rather than human transcription. These technological advances in transcription, along with shifts in the way that research is undertaken (for example, increasingly via video conferencing as a result of the COVID-19 pandemic), mean that the need to critically reflect upon the place of transcription in third sector research is more urgent.
In this article, I explore the place of transcription in qualitative research, with a focus on the importance of this process for third sector researchers. The article is structured as follows. First, I review the qualitative methods literature on audio transcription and the key themes that arise. Next, I report on a review undertaken of recent qualitative research articles in Voluntas and the way that authors discuss transcription in these articles. Finally, I propose a framework for qualitative third sector researchers to include transcription as part of their research design and elements to consider in including descriptions of the transcription process in writing up qualitative research.
Audio Transcription: What We Know
At a basic level, transcription refers to the transformation of recorded audio (usually spoken word) into a written form that can be used to analyze a particular phenomenon or event (Duranti, 2006 ). For many qualitative researchers, transcription has become a fairly taken-for-granted aspect of the research process. In this section, I review the methods literature on the process of audio (and video) transcription as part of qualitative research on the third sector, focusing on three key areas—how transcription is undertaken, epistemological and ethical considerations, and the role of technology.
Qualitative research and transcription
While quantitative research seeks to explain, generalize and predict patterns through the analysis of variables, qualitative research questions are more interested in understanding and interpreting the socially constructed world around us (Bryman, 2016 ). This means that data are collected through documents, observation and interviews, and the latter are often recorded in order to analyze these as documents. For third sector research, recordings are most commonly made of interviews and focus groups, but may also be of meetings, events and other activities to ensure that researchers do not have to rely on their power of recall or scribbled notes.
Transcription is a notoriously time-consuming and often tedious task which can take between three hours and over eight hours to transcribe one hour of audio, depending on typing speed. Transcription is not, however, a mechanical process where the written document becomes an objective record of the event—indeed, written text varies from the spoken word in terms of syntax, word choice and accepted grammar (Davidson, 2009 ). The transcriber therefore has to make subjective decisions throughout about what to include (or not), whether to correct mistakes and edit grammar and repetitions. This has been described as a spectrum between “naturalized” transcription (or “intelligent verbatim”) which adapts the oral to written norms, and “denaturalized” transcription (“full verbatim”), where everything is left in, including utterances, mistakes, repetitions and all grammatical errors (Bucholtz, 2000 ).
While some contend that denaturalized transcription is more ‘accurate’, the same can equally be argued for naturalized, as it allows the transcriber to omit occasions when, for instance, an individual mis-speaks and corrects themselves, thereby allowing the transcriber to record closer to what was intended and how the interviewee might have portrayed themselves in a written form. As Lapadat ( 2000 , p. 206) explains, “Spoken language is structured and accomplished differently than written text, so when talk is re-presented as written text, it is not surprising that readers draw on their knowledge of written language to evaluate it.” Other nonverbal cues, such as laughter, tone of voice (e.g. sarcasm, frustration, emphasis) and the use or omission of punctuation, can also drastically alter the meaning or intention of what an individual says. In addition, the transcriber must make decisions about how much contextual information to include, such as interruptions, crosstalk and inaudible segments (Lapadat, 2000 ). Because of the range of types of research that employ qualitative methods, there is no single set of rules for transcription but rather these decisions must be based on the research questions and approach.
Epistemological and Ethical Considerations
Because the researcher (or external transcriber) must make these decisions as they translate audio into written text, transcription is an inherently interpretative and political act, influenced by the transcriber’s own assumptions and biases (Jaffe, 2007 ). Every choice that the transcriber makes therefore shapes how the research participant is portrayed and determines what knowledge or information is relevant and valuable and what is not. Indeed, two transcribers may hear differently and select relevant spoken material differently (Stelma & Cameron, 2007 ). As Davidson ( 2009 ) notes (and as I explore in further detail in the next section), despite being a highly interpretive process, transcription is frequently depicted using positivist norms of knowledge creation.
Transcription also involves potential ethical considerations and dilemmas. When working with disadvantaged communities, deciding how to depict research participants in written text can highlight the challenges of ethical representation. As Kvale ( 1996 , pp. 172–3) notes, “Be mindful that the publication of incoherent and repetitive verbatim interview transcripts may involve an unethical stigmatization of specific persons or groups of people”. Oliver et al. ( 2005 ) similarly demonstrate how transcribers must make decisions about how to represent participants’ use of slang, colloquialisms and accents in ways that are accurate but also respectful of the respondent’s intended meaning. Some researchers decide to send finished transcriptions to interviewees for approval in order to honor commitments to fully informed consent, to ensure transcription accuracy or in some cases as a means to address the balance of power between the researcher and interviewee. As Mero-Jaffe ( 2011 ) describes, on the one hand, this may empower interviewees to control the way that they are portrayed in the research. On the other hand, Mero-Jaffe found that seeking transcript approval from interviewees sometimes increased their embarrassment at the way that their statements appear in text. This may be especially problematic with full verbatim transcriptions.
Technology and Transcription
As technology improves and AI becomes increasingly able to create written text from recorded audio, researchers might ask—is human transcription even necessary? New options in Computer Assisted Qualitative Data Analysis Software (CAQDAS) such as NVivo, Atlas.ti and MAXQDA give qualitative researchers the option to forgo audio-to-text transcription altogether, and instead engage in live coding of audio or video files. Using this method, researchers first watch or listen to recordings to code for nonverbal cues, followed by a stage of note taking and coding based on pre-defined themes and matching these with time codes and nonverbal cues. Finally, researchers then transcribe specific quotes of interest from the recording (Parameswaran et al., 2020 ). This process may improve immersion in the data and allow researchers to account for dynamics that are often lost in complete audio-to-text transcription, such as group interactions and nonverbal communication.
There is a considerable need to develop the evidence base on the role of AI in transcription for qualitative research, with many important publications that consider the issue (e.g. Gibbs et al., 2002 ; Markle et al., 2011 ) out-of-date given the swift rate of change in AI technologies. Over the last few years, voice and speech recognition technologies have improved dramatically and may now be able to provide researchers with “good enough” first drafts of transcripts (Bokhove & Downey, 2018 ), providing certain conditions are in place (e.g. limited number of speakers and excellent audio quality). Using these technologies can save researchers time and money. As a result of the COVID-19 pandemic, many qualitative researchers are now undertaking interviews over Zoom or other video conferencing apps, which is a trend that may continue beyond the pandemic (Dodds & Hess, 2020 ). Zoom offers AI live transcription options, which benefits from the generally clear audio quality of a video conference, compared to in-person interviews where there is a greater chance of audio interference and background noise that may be undetected in the moment.
While AI may offer a cheaper and quicker alternative to human transcription, these transcripts will need to be meticulously checked by the researcher to ensure accuracy, fill in missing details or edit for context and readability. Using cloud-based AI transcription services also raises potential ethical concerns about data protection and confidentiality (Da Silva, 2021 ). There are numerous subjective decisions made in the course of creating a transcription that AI is unable to process, such as where to include punctuation, which words to include or exclude (such as filler words, hesitations, etc.) and how to denote things such as interruptions, hesitations and nonverbal cues. Voice-to-text software is also generally less accurate in discerning multiple voices or different accents (Bokhove & Downey, 2018 ). Several studies have considered how researchers/transcribers can use voice recognition software to listen and repeat the spoken text of an interview into software as a shortcut to traditional typing transcription (Matheson, 2007 ; Tilley, 2003 ), but the above shortcomings and cautions apply.
Transcription and Third Sector Research
Transcription matters for third sector research because qualitative research methodologies make up a large percentage of studies undertaken on nonprofits—as much as 40–80% of research published in this field (Igalla et al., 2019 ; Laurett & Ferreira, 2018 ; von Schnurbein et al., 2018 ). Audio transcription is particularly important for third sector research for several reasons. In conducting qualitative research (which aims to produce rich, rigorous description) and as third sector researchers (who study organizations that seek to improve society and who may be working with traditionally disenfranchised or disadvantaged communities), we have a particular ethical obligation to ensure that our research provides an accurate depiction of our participants’ lives and the organizations with which they are involved.
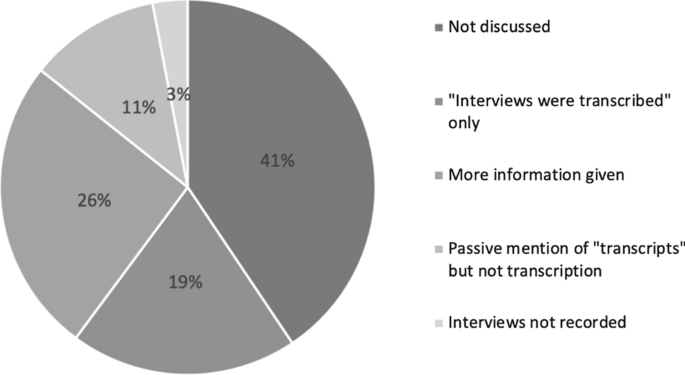
However, transcription is perhaps the most underacknowledged aspect of the qualitative research process, and this is also evident in the way that transcription is discussed in research articles. In order to survey the current depiction of the transcription process in third sector research, I undertook a review of the 212 most recent papers in Voluntas that include the word ‘interview’ to explore how qualitative research articles discuss transcription as part of their methodology. Footnote 1 Of these papers, 79 were deemed not applicable (because they were quantitative research papers that mentioned interviews in another context, or used the word interview to denote the administering of a structured questionnaire, or systematic review papers reporting on other research). This left 133 articles which were analyzed to explore the extent to which transcription was described—if at all—as part of the research methodology. Footnote 2
The analysis (illustrated in Fig. 1 ) found that 41% of papers employing interviews as a research method did not mention transcription at all, while 11% mentioned transcripts but not the process of transcription. It was not clear from these whether or not interviews were recorded or if researchers relied upon written notes taken during interviews, or how information from the oral interview was converted into analyzable text. The most common discussion of transcription (19%) was a simple sentence along the lines of “interviews were recorded and transcribed”, while 26% gave some further information including who undertook the transcription (the researcher(s), a research assistant or a commercial company) or that the interviews were transcribed ‘verbatim’ (with none explaining what they mean by this term). These findings are not dissimilar to a study of qualitative research in nursing, where it was found that 66% of articles reporting solely that interviews were transcribed, and the remaining articles indicated only “full” or “verbatim” to clarify the process (Wellard & McKenna, 2001 ). I also surveyed the first authors’ departmental affiliations/field of study to gauge any differences between academic fields (Table 1 ) although there were not considerable differences.
Transcription in Voluntas qualitative articles
The fact that over half of the Voluntas articles using interviews as a research method make no mention of the transcription process is a problem for transparency in qualitative research. This tendency may be a symptom of the fact that qualitative researchers face greater challenges in academic publishing that disadvantage longer from, in-depth qualitative research to fit within prescribed word limits (Moravcsik, 2014 ). In researchers’ efforts to ensure that qualitative research meets requirements for transparency, rigor and reliability, efforts are concentrated on descriptions of case and participant selection and data analysis while transcription as the conduit between data collection and analysis remains unproblematized. This emphasis reflects the growing influence of positivist views of validity. Ignoring the subjective decisions and theoretical perspectives that determine the creation of a transcript therefore inadvertently presupposes a positivist stance on the objective nature of data which is inconsistent with qualitative methodologies.
A Framework for Undertaking and Reporting on Transcription
As shown in the previous section, there is currently widespread neglect of transcription as part of interpretive qualitative research on the third sector. In this section, I present key elements for third sector researchers to consider in regard to transcription, both to ensure rigor as part of the qualitative research process and in writing up qualitative research, drawing upon examples of good practice from previous research in Voluntas. These recommendations are based on a review of the literature as well as my personal experience as a qualitative researcher, qualitative methods teacher, and professional transcriber.
Before Transcribing: Ethics and Data Management
All decisions regarding research design, data collection and data management should be made at the beginning of a qualitative research project when applying for ethical/IRB approval from one’s university, and this includes transcription. At this stage, the researcher should confirm with their university whether they have a budget for transcription. Undertaking ethical qualitative research means ensuring standards of transparency, informed consent, confidentiality and protection of the data obtained from the research (Blaxter et al., 2001 ). Increasing concerns about data protection and legislation such as GDPR in the European Union have prompted many universities to institute strict rules about where research data can be stored. Some universities do not allow the use of certain cloud servers, such as Dropbox. These considerations should be taken into account when deciding how to undertake and record interviews (Da Silva, 2021 )—for instance, if you are recording using your mobile phone, it is important to be sure you know whether recordings automatically upload to the cloud. For this reason, it may be preferable to use a traditional digital recorder so you can manually download the files to your computer and know exactly where everything is saved.
Before Transcribing: The Interview
Before transcription can even be considered, researchers must ensure that they have a suitable audio recording, which begins with the interview itself—whenever possible, interviews should be conducted in a quiet environment without background noise or interruptions and the audio recording device should be placed close enough to the respondent to pick up their voice clearly. While recording interviews with a mobile phone has become increasingly common and easy, using a backup recording device is always a good idea to mitigate against flat batteries, full memory cards, and human error. If recording with your mobile phone, it’s also critical to remember to place it on airplane mode/‘do not disturb’ for the duration of the interview.
To Transcribe or Not to Transcribe?
While transcription from audio recordings is considered standard practice in qualitative research (Tracy, 2019 ), it is not the only way of undertaking qualitative interviews, and it is important to note that there are many reasons why it may not be desirable, appropriate or possible to record interviews at all. In relation to third sector research, this is most commonly the case in community-based research, research with political elites or research in challenging environments. One article explained that they did not record interviews because: “In sectors marked by fear, intimidation, and strong security apparatuses, recording devices would almost certainly have led to self-censorship and limited our access.” (Atia & Herrold, 2018 , p. 1046). Similarly, researchers may be unable to record in community settings because of sub-optimal recording conditions (e.g. meeting outside, noisy environments, etc.) or because using recording device makes participants uncomfortable or reinforces power relations between the researcher and participants (Quintanilha et al., 2015 ).
If researchers decide not to comprehensively transcribe recordings, or decide not to record qualitative fieldwork at all, this should be noted and explained in relation to methods. Other methods of notetaking and analysis may be more suited to certain types of ethnographic research, such as reflexive journaling (Halcomb & Davidson, 2006 ), or Systematic and Reflexive Interviewing and Reporting—a process by which a researcher and research assistant jointly interview participants and write their own reports that include observations and analyses, which are collaboratively analyzed (Loubere, 2017 ).
How to Transcribe?
Traditionally, transcribers used foot pedals to play, rewind and fast forward tape recordings while they typed. Now that audio files are digital, several free and low cost programs are available (such as Express Scribe and oTranscribe) that let transcribers set up hot keys to perform the same actions without having to navigate away from their transcript document.
The degree of detail to include in transcripts should be decided upon before interviews are transcribed. This is important because previous research has demonstrated that the format selected for transcription significantly impacts how the researcher interprets the data (Mishler, 2003 ; Packer, 2017 ). There is no one best or “most accurate” style of transcription, but rather, a researcher should consider the particular theoretical background and research questions of the study in order to determine where on the scale of full verbatim to intelligent verbatim is most appropriate for the study. Because third sector research is most commonly associated with social science and business disciplines rather than linguistics, it will rarely be necessary or appropriate to employ the conventions of conversation analysis or extreme levels of denaturalized transcription (Bucholtz, 2000 ). Indeed, it might most frequently be appropriate to employ a version of naturalized/intelligent verbatim, so that any participants’ quotes included in written works are more ‘readable’ and do not include excessive repetitions or verbal fillers such as ‘um’.
If the researcher determines that naturalized or intelligent verbatim transcription is the most appropriate for their study, several considerations should be heeded in order to ensure that meaning is not distorted or lost. First, indications of laughter, nonverbal cues (such as sighs, huffs, finger-snaps, sobbing or even blowing raspberries) should be included if these convey important meaning. Other considerations of how to transcribe may be based more on personal preference and the ability to produce a document that is easily analyzable in the researcher’s chosen medium. For instance, wide margins on one side can be useful for researchers who choose to analyze their data on paper or in Microsoft Word, while other more flowing templates will work better to import into software such as NVivo. It can also be useful to include time stamps for unclear or inaudible statements, or at regular intervals (e.g. every minute) which makes it much easier to check a transcript against the original audio.
Who Transcribes?
As discussed in the consideration of qualitative studies, the prevalence of the passive voice when reporting on transcription (i.e. “interviews were transcribed”) obscures the important distinction of who undertook the transcription. If the researcher transcribes recordings themselves, then it is generally acceptable to assume the coherence between the research approach and approach to transcription, as well as the researcher’s confidence that the written transcript is an accurate record of the event/interview that took place. If, however, the researchers choose to outsource transcription to a research assistant or commercial transcription company, then care should be taken to give detailed and thorough instructions about the elements described above. The researcher should also spot check transcripts for accuracy, fill in any missed words/inaudibles and ensure that the transcription document fulfils their expectations in regard to level of verbatim, style and formatting.
Ideally, transcribers should be hired who have specialist knowledge of the subject matter and familiarity with the accents or dialect of the speakers. They should be provided with a key information about the project, such as the research questions, important terms and acronyms. Lapadat ( 2000 ) provides several useful suggestions when hiring transcribers in order to ensure transcription quality and increase rigor. First, rather than fully outsourcing transcription, the researchers can transcribe some interviews or portions of interviews themselves in order to provide an example for transcribers and develop a transcription protocol. Another option when employing research assistants to transcribe interviews is to include them directly in the interviews (either as a co-interviewer or observer), so they have direct involvement in the research and context.
Finally, when working with external transcribers it can also be valuable to encourage transcribers to keep memos of the transcription process or contextual observations and impressions that may not come through in the written text. For instance, does the interviewee sound tired, frustrated, distracted or nervous? Does the interviewer interrupt the respondent frequently (which the transcriber may choose to edit for readability)? Or did the interview take place somewhere public, like a cafe, which may have made the respondent more guarded? Such information is often lost, particularly in projects that involve multiple research team members (for instance, a PI, multiple interviewers, research assistants and/or professional transcribers).
Writing about Transcription
Due to limited space or word limits, it is not typically possible or desirable to include all of the above details in research articles. Instead, at a minimum, researchers should include who transcribed the audio recordings as part of a commitment to ethical and transparent qualitative research. If this was done by anyone other than the researchers, authors should ideally describe the measures taken to ensure accuracy (developing a protocol for transcribers, spot checking, proofreading, sending transcripts to interviewees if appropriate) and ethical considerations (such as data protection and confidentiality).
Second, researchers should indicate the type of transcription—whether selective (pulling out relevant quotes and themes, or transcribing just the ‘gist’), intelligent verbatim/naturalized or full verbatim/denaturalized. The choice of type of transcription should align to the researcher’s epistemological position and theoretical framework.
Finally, researchers should include any other subjective decision-making that took place during the transcription process, in much the same way that researchers are encouraged to be transparent about their subjectivity and positionality in undertaking interviews and analysis of qualitative data (McCorkel & Myers, 2003 ). This may include information about selecting the level of verbatim, working with external transcribers, feedback from interviewees on transcripts or efforts to ensure accuracy of transcripts and coherence with the research approach.
The following quotes provide good examples of how to write about transcription:
The interviews, which were conducted in the native language of the interviewees by six female Hebrew-Arabic-speaking interviewers, were recorded, translated, and transcribed verbatim. […] Immediately following the interview, each interviewer transcribed and translated her interviews into Hebrew. In this manner, we sought to achieve a translation that was as close as possible to the interviewer’s insights regarding the participants, and we regarded the interviewers as active agents in the creation of knowledge. (Yanay-Ventura et al., 2020 , p. 6) Three Spanish speaking investigators transcribed all of the interviews from audio recording devices, checked each other’s transcription for accuracy, and analyzed the interviews using thematic analysis (Braun & Clarke, 2006 ). The transcribers observed the focus groups and took notes on participants’ voices and other identifying traits to help the transcription process go more smoothly. Researchers aided the transcribers in this regard by asking participants at the beginning of the focus groups to introduce themselves using a pseudonym and briefly remark upon how they preferred to spend their time. (Schwingel et al., 2017 , p. 170)
In both of these examples, the authors treat the process of transcription as part of the broader research process, rather than as an automatic conversion of audio to text. While there is limited clarification about the type of transcription (beyond ‘verbatim’), the discussion of the subjective decision-making as part of the transcription process and acknowledgment of the agency of the individuals undertaking transcription increases transparency and therefore rigor.
Conclusions
Qualitative research can help us to understand some of the important issues impacting the third sector in ways that quantitative methods fall short of explaining, such as the ways that individuals and organizations make sense of public policy and societal challenges, how and why organizations design their services and activities in particular ways, and the intricacies of the relationships between boards, executives, staff and volunteers. Qualitative methods training stresses that an interpretivist epistemological position sees knowledge as socially constructed, yet transcription has slipped through the cracks of methodological examination in the process of creating and interpreting meaning.
In this short article, I sought to draw our attention to this important stage of qualitative data collection and analysis and call on third sector researchers to critically reflect upon transcription both in conducting research and in writing about it. I have focused primarily on the transcription of interviews , rather than focus groups or other multi-person events. All of the points raised in my framework transcription apply to these methods of data collection as well; however, there are further issues that need to be taken into consideration regarding focus groups that warrant further attention, such as the issues of power and accuracy of transcription when there are multiple people speaking and interrupting one another. Researchers employing multi-person recordings should therefore devote more time and consideration to transcription. Finally, technology continues to advance in the area of voice recognition, which may save researchers considerable time and/or money in transcription; however, I implore scholars to see transcription through an interpretivist rather than positivist lens, to ensure that the production of written transcripts is not approached as the creation of objective knowledge.
While this approach may have obscured other methods that employ transcription, such as focus groups, the intention of the survey is to provide a snapshot illustration of transcription and qualitative methods rather than a systematic review.
Articles reviewed are listed in Appendix 1.
Atia, M., & Herrold, C. E. (2018). Governing through patronage: The rise of NGOs and the fall of civil society in Palestine and Morocco. VOLUNTAS: International Journal of Voluntary and Nonprofit Organizations, 29 (5), 1044–1054. https://doi.org/10.1007/s11266-018-9953-6
Article Google Scholar
Blaxter, L., Hughes, C., & Tight, M. (2001). How to research (2nd ed.). Open University Press.
Bokhove, C., & Downey, C. (2018). Automated generation of “good enough” transcripts as a first step to transcription of audio-recorded data. Methodological Innovations , 11 (2). https://doi.org/10.1177/2059799118790743 .
Braun, V., & Clarke, V. (2006). Using thematic analysis in psychology. Qualitative Research in Psychology , 3 (2), 77–101. https://doi.org/10.1191/1478088706qp063oa .
Bryman, A. (2016). Social research methods (5th ed.). Oxford University Press.
Bucholtz, M. (2000). The politics of transcription. Journal of Pragmatics, 32 (10), 1439–1465. https://doi.org/10.1016/S0378-2166(99)00094-6
Da Silva, J. (2021). Producing ‘good enough’ automated transcripts securely: Extending Bokhove and Downey (2018) to address security concerns. Methodological Innovations, 14 (1), 2059799120987766. https://doi.org/10.1177/2059799120987766
Davidson, C. (2009). Transcription: Imperatives for qualitative research. International Journal of Qualitative Methods, 8 (2), 35–52.
Dodds, S., & Hess, A. C. (2020). Adapting research methodology during COVID-19: Lessons for transformative service research. Journal of Service Management, 32 (2), 203–217. https://doi.org/10.1108/JOSM-05-2020-0153
Duranti, A. (2006). Transcripts, like shadows on a wall. Mind, Culture, and Activity, 13 (4), 301–310. https://doi.org/10.1207/s15327884mca1304_3
Gibbs, G. R., Friese, S., & Mangabeira, W. C. (2002). View of the use of new technology in qualitative research. Orum Qualitative Sozialforschung/Forum: Qualitative Social Research , 3 (2). https://www.qualitative-research.net/index.php/fqs/article/view/847/1840
Halcomb, E. J., & Davidson, P. M. (2006). Is verbatim transcription of interview data always necessary? Applied Nursing Research, 19 (1), 38–42. https://doi.org/10.1016/j.apnr.2005.06.001
Igalla, M., Edelenbos, J., & van Meerkerk, I. (2019). Citizens in action, what do they accomplish? A systematic literature review of citizen initiatives, their main characteristics, outcomes, and factors. VOLUNTAS: International Journal of Voluntary and Nonprofit Organizations, 30 (5), 1176–1194. https://doi.org/10.1007/s11266-019-00129-0
Jaffe, A. (2007). Variability in transcription and the complexities of representation, authority and voice. Discourse Studies, 9 (6), 831–836. https://doi.org/10.1177/1461445607082584
Kvale, S. (1996). Interviews: An introduction to qualitative research interviewing . Sage.
Lapadat, J. C. (2000). Problematizing transcription: Purpose, paradigm and quality. International Journal of Social Research Methodology, 3 (3), 203–219. https://doi.org/10.1080/13645570050083698
Laurett, R., & Ferreira, J. J. (2018). Strategy in nonprofit organisations: A systematic literature review and agenda for future research. VOLUNTAS: International Journal of Voluntary and Nonprofit Organizations, 29 (5), 881–897. https://doi.org/10.1007/s11266-017-9933-2
Loubere, N. (2017). Questioning transcription: The case for the systematic and reflexive interviewing and reporting (SRIR) method. Forum Qualitative Sozialforschung/Forum: Qualitative Social Research , 18 (2), Article 2. https://doi.org/10.17169/fqs-18.2.2739
Markle, D. T., West, R. E., & Rich, P. J. (2011). Beyond transcription: Technology, change, and refinement of method. Forum Qualitative Sozialforschung/Forum: Qualitative Social Research , 12 (3), Article 3. https://doi.org/10.17169/fqs-12.3.1564
Matheson, J. L. (2007). The voice transcription technique: Use of voice recognition software to transcribe digital interview data in qualitative research. Qualitative Report, 12 (4), 547–560.
Google Scholar
McCorkel, J. A., & Myers, K. (2003). What difference does difference make? Position and privilege in the field. Qualitative Sociology, 26 (2), 199–231. https://doi.org/10.1023/A:1022967012774
Mero-Jaffe, I. (2011). ‘Is that what I Said?’ Interview transcript approval by participants: An aspect of ethics in qualitative research. International Journal of Qualitative Methods, 10 (3), 231–247. https://doi.org/10.1177/160940691101000304
Mishler, E. (2003). Representing discourse: The rhetoric of transcription. In N. Fielding (Ed.), Interviewing . Sage.
Moravcsik, A. (2014). Transparency: The revolution in qualitative research. PS: Political Science & Politics , 47 (1), 48–53. https://doi.org/10.1017/S1049096513001789
Oliver, D. G., Serovich, J. M., & Mason, T. L. (2005). Constraints and opportunities with interview transcription: Towards reflection in qualitative research. Social Forces, 84 (2), 1273–1289. https://doi.org/10.1353/sof.2006.0023
Packer, M. J. (2017). The science of qualitative research . Cambridge University Press.
Book Google Scholar
Parameswaran, U. D., Ozawa-Kirk, J. L., & Latendresse, G. (2020). To live (code) or to not: A new method for coding in qualitative research. Qualitative Social Work, 19 (4), 630–644. https://doi.org/10.1177/1473325019840394
Quintanilha, M., Mayan, M. J., Thompson, J., & Bell, R. C. (2015). Different approaches to cross-lingual focus groups: Lessons from a cross-cultural community-based participatory research project in the ENRICH study. International Journal of Qualitative Methods, 14 (5), 1609406915621419. https://doi.org/10.1177/1609406915621419
Schwingel, A., Wiley, A., Teran-Garcia, M., McCaffrey, J., Gálvez, P., Hawn, R., Villegas, E., Coba, S., Vizcarra, M., Luty, G., Montez, R., & The Abriendo Caminos Promotora Project Group. (2017). More than help? Volunteerism in US Latino culture. VOLUNTAS: International Journal of Voluntary and Nonprofit Organizations , 28 (1), 162–183. https://doi.org/10.1007/s11266-016-9731-2
Stelma, J. H., & Cameron, L. J. (2007). Intonation units in spoken interaction: Developing transcription skills. Text & Talk, 27 (3), 361–393. https://doi.org/10.1515/TEXT.2007.015
Tilley, S. A. (2003). “Challenging” research practices: Turning a critical lens on the work of transcription. Qualitative Inquiry, 9 (5), 750–773. https://doi.org/10.1177/1077800403255296
Tracy, S. J. (2019). Qualitative research methods: Collecting evidence, crafting analysis (2nd ed.). John Wiley & Sons.
von Schnurbein, G., Perez, M., & Gehringer, T. (2018). Nonprofit comparative research: Recent agendas and future trends. VOLUNTAS: International Journal of Voluntary and Nonprofit Organizations, 29 (3), 437–453. https://doi.org/10.1007/s11266-017-9877-6
Wellard, S., & McKenna, L. (2001). Turning tapes into text: Issues surrounding the transcription of interviews. Contemporary Nurse, 11 (2–3), 180–186. https://doi.org/10.5172/conu.11.2-3.180
Yanay-Ventura, G., Issaq, L., & Sharabi, M. (2020). Civic service and social class: The case of young Arab women in Israel. VOLUNTAS: International Journal of Voluntary and Nonprofit Organizations . https://doi.org/10.1007/s11266-020-00210-z
Download references
No funding was received to assist with the preparation of this manuscript.
Author information
Authors and affiliations.
Department of Politics & Society, Aalborg University, Aalborg, Denmark
Caitlin McMullin
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Caitlin McMullin .
Ethics declarations
Conflict of interest.
The author declares that they have no conflict of interest.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix 1: Articles Reviewed
Aasland, A., Kropp, S., & Meylakhs, A. Y. (2020). Between Collaboration and Subordination: State and Non-state Actors in Russian Anti-drug Policy. Voluntas , 31 (2), 422–436. https://doi.org/10.1007/s11266-019-00158-9
Åberg, P., Einarsson, S., & Reuter, M. (2021). Think Tanks: New Organizational Actors in a Changing Swedish Civil Society. Voluntas , 32 (3), 634–648. https://doi.org/10.1007/s11266-019-00174-9
Afkhami, A., Nasr Isfahani, A., Abzari, M., & Teimouri, H. (2019). Toward a Deep Insight into Employee Participation in Employer-Supported Volunteering in Iranian Organizations: A Grounded Theory. Voluntas , 30 (5), 1036–1053. https://doi.org/10.1007/s11266-019-00141-4
Anasti, T. (2020). The Strategic Action Field of Sex Work and Sex Trafficking: A Case Study of a Contentious Field in Chicago. Voluntas , 31 (1), 169–183. https://doi.org/10.1007/s11266-019-00161-0
Appe, S. (2018). Directions in a Post-aid World? South–South Development Cooperation and CSOs in Latin America. Voluntas , 29 (2), 271–283. https://doi.org/10.1007/s11266-017-9838-0
Arhin, A. A., Kumi, E., & Adam, M.-A. S. (2018). Facing the Bullet? Non-Governmental Organisations’ (NGOs’) Responses to the Changing Aid Landscape in Ghana. Voluntas , 29 (2), 348–360. https://doi.org/10.1007/s11266-018-9966-1
Arvidson, M. (2018). Change and Tensions in Non-profit Organizations: Beyond the Isomorphism Trajectory. Voluntas , 29 (5), 898–910. https://doi.org/10.1007/s11266-018-0021-z
Atia, M., & Herrold, C. E. (2018). Governing Through Patronage: The Rise of NGOs and the Fall of Civil Society in Palestine and Morocco. Voluntas , 29 (5), 1044–1054. https://doi.org/10.1007/s11266-018-9953-6
Ávila, L., & Amorim, M. (2021). Organisational Identity of Social Enterprises: A Taxonomic Approach. Voluntas , 32 (1), 13–27. https://doi.org/10.1007/s11266-020-00264-z
Baillie Smith, M., Fadel, B., O’Loghlen, A., & Hazeldine, S. (2020). Volunteering Hierarchies in the Global South: Remuneration and Livelihoods. Voluntas . https://doi.org/10.1007/s11266-020-00254-1
Bandini, F., Gigli, S., & Mariani, L. (2021). Social Enterprises and Public Value: A Multiple-Case Study Assessment. Voluntas , 32 (1), 61–77. https://doi.org/10.1007/s11266-020-00285-8
Bano, M. (2019). Partnerships and the Good-Governance Agenda: Improving Service Delivery Through State–NGO Collaborations. Voluntas , 30 (6), 1270–1283. https://doi.org/10.1007/s11266-017-9937-y
Barinaga, E. (2020). Coopted! Mission Drift in a Social Venture Engaged in a Cross-Sectoral Partnership. Voluntas , 31 (2), 437–449. https://doi.org/10.1007/s11266-018-0019-6
Bayalieva-Jailobaeva, K. (2018). New Donor Strategies: Implications for NGOs in Post-Soviet Kyrgyzstan. Voluntas , 29 (2), 284–295. https://doi.org/10.1007/s11266-017-9878-5
Beaton, E. E. (2021). No Margin, No Mission: How Practitioners Justify Nonprofit Managerialization. Voluntas , 32 (3), 695–708. https://doi.org/10.1007/s11266-019-00189-2
Bergfeld, A., Plagmann, C., & Lutz, E. (2021). Know Your Counterparts: The Importance of Wording for Stakeholder Communication in Social Franchise Enterprises. Voluntas , 32 (1), 104–119. https://doi.org/10.1007/s11266-020-00289-4
Bidet, E., Eum, H., & Ryu, J. (2018). Diversity of Social Enterprise Models in South Korea. Voluntas , 29 (6), 1261–1273. https://doi.org/10.1007/s11266-018-9951-8
Bies, A., & Kennedy, S. (2019). The State and the State of the Art on Philanthropy in China. Voluntas , 30 (4), 619–633. https://doi.org/10.1007/s11266-019-00142-3
Bradford, A., Luke, B., & Furneaux, C. (2020). Exploring Accountability in Social Enterprise: Priorities, Practicalities, and Legitimacy. Voluntas , 31 (3), 614–626. https://doi.org/10.1007/s11266-020-00215-8
Cannon, S. M. (2020). Legitimacy as Property and Process: The Case of an Irish LGBT Organization. Voluntas , 31 (1), 39–55. https://doi.org/10.1007/s11266-019-00091-x
Carlsen, H. B., Doerr, N., & Toubøl, J. (2020). Inequality in Interaction: Equalising the Helper–Recipient Relationship in the Refugee Solidarity Movement. Voluntas . https://doi.org/10.1007/s11266-020-00268-9
Chatterjee, D., Subramanian, B., & Hota, P. K. (2020). Professionalization and Hybridization Dynamics of Social Enterprises. Voluntas , 31 (3), 457–471. https://doi.org/10.1007/s11266-020-00217-6
Chatzichristos, G., & Nagopoulos, N. (2020). Social Entrepreneurship and Institutional Sustainability: Insights from an Embedded Social Enterprise. Voluntas , 31 (3), 484–493. https://doi.org/10.1007/s11266-019-00188-3
Chaves-Avila, R., & Savall-Morera, T. (2019). The Social Economy in a Context of Austerity Policies: The Tension Between Political Discourse and Implemented Policies in Spain. Voluntas , 30 (3), 487–498. https://doi.org/10.1007/s11266-018-00075-3
Chewinski, M. (2019). Coordinating Action: NGOs and Grassroots Groups Challenging Canadian Resource Extraction Abroad. Voluntas , 30 (2), 356–368. https://doi.org/10.1007/s11266-018-0023-x
Clear, A., Paull, M., & Holloway, D. (2018). Nonprofit Advocacy Tactics: Thinking Inside The Box? Voluntas , 29 (4), 857–869. https://doi.org/10.1007/s11266-017-9907-4
Clerkin, B., & Quinn, M. (2019). Restricted Funding: Restricting Development? Voluntas , 30 (6), 1348–1364. https://doi.org/10.1007/s11266-018-00048-6
Cookingham Bailey, E. (2020). Advocacy and Service Delivery in the Voluntary Sector: Exploring the History of Voluntary Sector Activities for New Minority and Migrant Groups in East London, 1970s–1990s. Voluntas . https://doi.org/10.1007/s11266-020-00253-2
Crack, A. M. (2018). The Regulation of International NGOS: Assessing the Effectiveness of the INGO Accountability Charter. Voluntas , 29 (2), 419–429. https://doi.org/10.1007/s11266-017-9866-9
Crotty, J., & Ljubownikow, S. (2020). Creating Organisational Strength from Operationalising Restrictions: Welfare Non-profit Organisations in the Russian Federation. Voluntas , 31 (6), 1148–1158. https://doi.org/10.1007/s11266-020-00271-0
Deng, G. (2019). Trends in Overseas Philanthropy by Chinese Foundations. Voluntas , 30 (4), 678–691. https://doi.org/10.1007/s11266-017-9868-7
Dinh, K., Hong, K. T., Haire, B., & Worth, H. (2021). Historic and Contemporary Influences on HIV Advocacy in Vietnam. Voluntas , 32 (3), 610–620. https://doi.org/10.1007/s11266-020-00220-x
Dong, Q., Guo, J., & Huang, C.-C. (2019). Nonprofit Alliance in China: Effects of Alliance Process on Goal Achievement. Voluntas , 30 (2), 300–311. https://doi.org/10.1007/s11266-018-9990-1
Edenfield, A. C., & Andersson, F. O. (2018). Growing Pains: The Transformative Journey from a Nascent to a Formal Not-For-Profit Venture. Voluntas , 29 (5), 1033–1043. https://doi.org/10.1007/s11266-017-9936-z
Elsayed, Y. (2018). At the Intersection of Social Entrepreneurship and Social Movements: The Case of Egypt and the Arab Spring. Voluntas , 29 (4), 819–831. https://doi.org/10.1007/s11266-017-9943-0
Eschweiler, J., Svensson, S., Mocca, E., Cartwright, A., & Villadsen Nielsen, L. (2019). The Reciprocity Dimension of Solidarity: Insights from Three European Countries. Voluntas , 30 (3), 549–561. https://doi.org/10.1007/s11266-018-0031-x
Eynaud, P., Juan, M., & Mourey, D. (2018). Participatory Art as a Social Practice of Commoning to Reinvent the Right to the City. Voluntas , 29 (4), 621–636. https://doi.org/10.1007/s11266-018-0006-y
Falkenhain, M. (2020). Dividing Lines: Understanding the Creation and Replication of Fragmentations Among NGOs in Hybrid Regimes. Voluntas , 31 (4), 663–673. https://doi.org/10.1007/s11266-019-00185-6
Fang, Q., Fisher, K. R., & Li, B. (2020). Follower or Challenger? How Chinese Non-governmental Organizations Manage Accountability Requirements from Funders. Voluntas , 31 (4), 722–735. https://doi.org/10.1007/s11266-019-00184-7
Farid, M., & Li, H. (2021). Reciprocal Engagement and NGO Policy Influence on the Local State in China. Voluntas , 32 (3), 597–609. https://doi.org/10.1007/s11266-020-00288-5
Fehsenfeld, M., & Levinsen, K. (2019). Taking Care of the Refugees: Exploring Advocacy and Cross-sector Collaboration in Service Provision for Refugees. Voluntas , 30 (2), 422–435. https://doi.org/10.1007/s11266-019-00097-5
Ferguson, G. (2018). The Social Economy in Bolivia: Indigeneity, Solidarity, and Alternatives to Capitalism. Voluntas , 29 (6), 1233–1243. https://doi.org/10.1007/s11266-018-0013-z
Fulton, B. R., & Wood, R. L. (2018). Civil Society Organizations and the Enduring Role of Religion in Promoting Democratic Engagement. Voluntas , 29 (5), 1068–1079. https://doi.org/10.1007/s11266-018-9965-2
Gaby, S. (2020). Reconfiguring Organizational Commitments: Boundary Crossing in Civic Groups. Voluntas , 31 (6), 1121–1133. https://doi.org/10.1007/s11266-020-00272-z
Gaeta, G. L., Ghinoi, S., Silvestri, F., & Trasciani, G. (2021). Exploring Networking of Third Sector Organizations: A Case Study Based on the Quartieri Spagnoli Neighborhood in Naples (Italy). Voluntas , 32 (4), 750–766. https://doi.org/10.1007/s11266-020-00241-6
García-Orosa, B., & Pérez-Seijo, S. (2020). The Use of 360° Video by International Humanitarian Aid Organizations to Spread Social Messages and Increase Engagement. Voluntas , 31 (6), 1311–1329. https://doi.org/10.1007/s11266-020-00280-z
Grazioli, M., & Caciagli, C. (2018). Resisting to the Neoliberal Urban Fabric: Housing Rights Movements and the Re-appropriation of the ‘Right to the City’ in Rome, Italy. Voluntas , 29 (4), 697–711. https://doi.org/10.1007/s11266-018-9977-y
Grubb, A., & Henriksen, L. S. (2019). On the Changing Civic Landscape in Denmark and its Consequences for Civic Action. Voluntas , 30 (1), 62–73. https://doi.org/10.1007/s11266-018-00054-8
Guha, P. (2019). Going to Scale: A Case Study of an Indian Educational NGO. Voluntas , 30 (6), 1365–1379. https://doi.org/10.1007/s11266-019-00099-3
Guo, C., & Lai, W. (2019). Community Foundations in China: In Search of Identity? Voluntas , 30 (4), 647–663. https://doi.org/10.1007/s11266-017-9932-3
Heckert, R., Boumans, J., & Vliegenthart, R. (2020). How to Nail the Multiple Identities of an Organization? A Content Analysis of Projected Identity. Voluntas , 31 (1), 129–141. https://doi.org/10.1007/s11266-019-00157-w
Heideman, L. J. (2018). Making Civil Society Sustainable: The Legacy of USAID in Croatia. Voluntas , 29 (2), 333–347. https://doi.org/10.1007/s11266-017-9896-3
Hjort, M., & Beswick, D. (2020). Volunteering and Policy Makers: The Political Uses of the UK Conservative Party’s International Development Volunteering Projects. Voluntas . https://doi.org/10.1007/s11266-020-00222-9
Holthaus, L. (2019). Furthering Pluralism? The German Foundations in Transitional Tunisia. Voluntas , 30 (6), 1284–1296. https://doi.org/10.1007/s11266-018-00074-4
Horvath, A. (2020). The Transformative Potential of Experience: Learning, Group Dynamics, and the Development of Civic Virtue in a Mobile Soup Kitchen. Voluntas , 31 (5), 981–994. https://doi.org/10.1007/s11266-018-00062-8
Horvath, A., Brandtner, C., & Powell, W. W. (2018). Serve or Conserve: Mission, Strategy, and Multi-Level Nonprofit Change During the Great Recession. Voluntas , 29 (5), 976–993. https://doi.org/10.1007/s11266-017-9948-8
Huang, Y. (2019). At the Mercy of the State: Health Philanthropy in China. Voluntas , 30 (4), 634–646. https://doi.org/10.1007/s11266-018-9960-7
Ismail, A., & Johnson, B. (2019). Managing Organizational Paradoxes in Social Enterprises: Case Studies from the MENA Region. Voluntas , 30 (3), 516–534. https://doi.org/10.1007/s11266-018-00083-3
Jezierska, K., & Polanska, D. V. (2018). Social Movements Seen as Radical Political Actors: The Case of the Polish Tenants’ Movement. Voluntas , 29 (4), 683–696. https://doi.org/10.1007/s11266-017-9917-2
Kampen, T., Veldboer, L., & Kleinhans, R. (2019). The Obligation to Volunteer as Fair Reciprocity? Welfare Recipients’ Perceptions of Giving Back to Society. Voluntas , 30 (5), 991–1005. https://doi.org/10.1007/s11266-018-00082-4
Kang, Y. (2019). What Does China’s Twin-Pillared NGO Funding Game Entail? Growing Diversity and Increasing Isomorphism. Voluntas , 30 (3), 499–515. https://doi.org/10.1007/s11266-018-00085-1
Kappelides, P., Cuskelly, G., & Hoye, R. (2019). The Influence of Volunteer Recruitment Practices and Expectations on the Development of Volunteers’ Psychological Contracts. Voluntas , 30 (1), 259–271. https://doi.org/10.1007/s11266-018-9986-x
Kennedy, D. (2019). The Inherently Contested Nature of Nongovernmental Accountability: The Case of HAP International. Voluntas , 30 (6), 1393–1405. https://doi.org/10.1007/s11266-019-00134-3
Kewes, A., & Munsch, C. (2019). Should I Stay or Should I Go? Engaging and Disengaging Experiences in Welfare-Sector Volunteering. Voluntas , 30 (5), 1090–1103. https://doi.org/10.1007/s11266-019-00122-7
Korstenbroek, T., & Smets, P. (2019). Developing the Potential for Change: Challenging Power Through Social Entrepreneurship in the Netherlands. Voluntas , 30 (3), 475–486. https://doi.org/10.1007/s11266-019-00107-6
Koubek, M. (2020). When Did They Protest? Beyond Co-optation or Channeling: Effects of EU Funding on Czech Romani NGOs and Civil Society. Voluntas , 31 (2), 404–421. https://doi.org/10.1007/s11266-018-0008-9
Kravchenko, Z., & Moskvina, A. (2018). Entrepreneurial NPOs in Russia: Rationalizing the Mission. Voluntas , 29 (5), 962–975. https://doi.org/10.1007/s11266-018-0016-9
Kumi, E. (2019). Aid Reduction and NGDOs’ Quest for Sustainability in Ghana: Can Philanthropic Institutions Serve as Alternative Resource Mobilisation Routes? Voluntas , 30 (6), 1332–1347. https://doi.org/10.1007/s11266-017-9931-4
Lall, S. A. (2019). From Legitimacy to Learning: How Impact Measurement Perceptions and Practices Evolve in Social Enterprise–Social Finance Organization Relationships. Voluntas , 30 (3), 562–577. https://doi.org/10.1007/s11266-018-00081-5
Lang, R., & Mullins, D. (2020). Field Emergence in Civil Society: A Theoretical Framework and Its Application to Community-Led Housing Organisations in England. Voluntas , 31 (1), 184–200. https://doi.org/10.1007/s11266-019-00138-z
Laurent, A., Garaudel, P., Schmidt, G., & Eynaud, P. (2020). Civil Society Meta-organizations and Legitimating Processes: The Case of the Addiction Field in France. Voluntas , 31 (1), 19–38. https://doi.org/10.1007/s11266-019-00094-8
Lee, E. K. M., & Chandra, Y. (2020). Dynamic and Marketing Capabilities as Predictors of Social Enterprises’ Performance. Voluntas , 31 (3), 587–600. https://doi.org/10.1007/s11266-019-00155-y
Li, S. (2020). Global Civil Society Under the New INGO Regulatory Law: A Comparative Case Study on Two INGOs in China. Voluntas , 31 (4), 751–761. https://doi.org/10.1007/s11266-019-00101-y
Li, S., & Wang, X. (2020). Seeking Credibility from Uncertainty: How Formal Cooptation Institution Unleashes Outspoken NGOs. Voluntas , 31 (4), 711–721. https://doi.org/10.1007/s11266-020-00204-x
Liu, F., & Zhang, X. (2020). Who are Volunteers in Urban China? Voluntas . https://doi.org/10.1007/s11266-020-00251-4
Lund, V., & Juujärvi, S. (2018). Residents’ Agency Makes a Difference in Volunteering in an Urban Neighbourhood. Voluntas , 29 (4), 756–769. https://doi.org/10.1007/s11266-018-9955-4
Marie, B., Isabelle, M., Jordan, B.-L., Hélène, C., Sophie, É., Julie, F., Christine, M., & Andrée, S. (2020). Promising Practices of Nonprofit Organizations to Respond to the Challenges Faced in Countering the Mistreatment of Older Adults. Voluntas , 31 (6), 1359–1370. https://doi.org/10.1007/s11266-020-00252-3
Martinez, D. E., & Cooper, D. J. (2020). Seeing Through the Logical Framework. Voluntas , 31 (6), 1239–1253. https://doi.org/10.1007/s11266-020-00223-8
Mathews, M. A. (2020). The Embeddedness of Nonprofit Leadership in Civic Governance. Voluntas , 31 (1), 201–212. https://doi.org/10.1007/s11266-019-00139-y
Mati, J. M. (2020). Civil Society in ‘Politics’ and ‘Development’ in African Hybrid Regimes: The Kenyan Case. Voluntas , 31 (4), 674–687. https://doi.org/10.1007/s11266-020-00211-y
Maya-Jariego, I., Holgado-Ramos, D., González-Tinoco, E., Muñoz-Alvis, A., & Ortega, M. (2020). More Money, More Problems? Resource Dependence and Professionalization of Non-governmental Social Services Organizations in Southern Spain. Voluntas , 31 (6), 1212–1225. https://doi.org/10.1007/s11266-020-00256-z
McLennan, B. J. (2020). Conditions for Effective Coproduction in Community-Led Disaster Risk Management. Voluntas , 31 (2), 316–332. https://doi.org/10.1007/s11266-018-9957-2
McMullin, C., & Skelcher, C. (2018). The Impact of Societal-Level Institutional Logics on Hybridity: Evidence from Nonprofit Organizations in England and France. Voluntas , 29 (5), 911–924. https://link.springer.com/article/10.1007/s11266-018-9996-8
Meijs, L., Handy, F., Simons, F.-J., & Roza, L. (2020). A Social Innovation: Addressing Relative Food Insecurity and Social Exclusion. Voluntas , 31 (5), 894–906. https://doi.org/10.1007/s11266-019-00105-8
Meyer, M., & Simsa, R. (2018). Organizing the Unexpected: How Civil Society Organizations Dealt with the Refugee Crisis. Voluntas , 29 (6), 1159–1175. https://doi.org/10.1007/s11266-018-00050-y
Mikołajczak, P. (2020). Social Enterprises’ Hybridity in the Concept of Institutional Logics: Evidence from Polish NGOs. Voluntas , 31 (3), 472–483. https://doi.org/10.1007/s11266-020-00195-9
Milbourn, B., Black, M. H., & Buchanan, A. (2019). Why People Leave Community Service Organizations: A Mixed Methods Study. Voluntas , 30 (1), 272–281. https://doi.org/10.1007/s11266-018-0005-z
Molina, J. L., Valenzuela-García, H., Lubbers, M. J., Escribano, P., & Lobato, M. M. (2018). “The Cowl Does Make The Monk”: Understanding the Emergence of Social Entrepreneurship in Times of Downturn. Voluntas , 29 (4), 725–739. https://doi.org/10.1007/s11266-017-9921-6
Moraes, R. L., & Andion, C. (2018). Civil Society and Social Innovation in Public Arenas in Brazil: Trajectory and Experience of the Movement Against Electoral Corruption (MCCE). Voluntas , 29 (4), 801–818. https://doi.org/10.1007/s11266-017-9867-8
Morais-da-Silva, R. L., Segatto, A. P., & Bezerra-de-Sousa, I. G. (2020). Connecting Two Sides: A Qualitative Study on Social Innovation Ventures and Poor Communities in an Emerging Economy. Voluntas , 31 (5), 966–980. https://doi.org/10.1007/s11266-019-00156-x
Musah-Surugu, I. J., Bawole, J. N., & Ahenkan, A. (2019). The “Third Sector” and Climate Change Adaptation Governance in Sub-Saharan Africa: Experience from Ghana. Voluntas , 30 (2), 312–326. https://doi.org/10.1007/s11266-018-9962-5
Mutongwizo, T. (2018). Comparing NGO Resilience and ‘Structures of Opportunity’ in South Africa and Zimbabwe (2010–2013). Voluntas , 29 (2), 373–387. https://doi.org/10.1007/s11266-017-9881-x
Näslund, H. (2020). Collective Deliberations and Hearts on Fire: Experiential Knowledge Among Entrepreneurs and Organisations in the Mental Health Service User Movement. Voluntas . https://doi.org/10.1007/s11266-020-00233-6
Nimu, A. (2018). Surviving Mechanisms and Strategies of Gender Equality NGOs in Romania and Poland. Voluntas , 29 (2), 310–332. https://doi.org/10.1007/s11266-018-9961-6
Noh, J.-E. (2019). Human Rights-Based Child Sponsorship: A Case Study of ActionAid. Voluntas , 30 (6), 1420–1432. https://doi.org/10.1007/s11266-018-0010-2
Obaze, Y. (2020). Supply Chain Challenges and Shared Value Destruction in the Community-Based Supply Chain. Voluntas , 31 (3), 550–562. https://doi.org/10.1007/s11266-020-00202-z
Onyx, J., Darcy, S., Grabowski, S., Green, J., & Maxwell, H. (2018). Researching the Social Impact of Arts and Disability: Applying a New Empirical Tool and Method. Voluntas , 29 (3), 574–589. https://doi.org/10.1007/s11266-018-9968-z
Pape, U., Brandsen, T., Pahl, J. B., Pieliński, B., Baturina, D., Brookes, N., Chaves-Ávila, R., Kendall, J., Matančević, J., Petrella, F., Rentzsch, C., Richez-Battesti, N., Savall-Morera, T., Simsa, R., & Zimmer, A. (2020). Changing Policy Environments in Europe and the Resilience of the Third Sector. Voluntas , 31 (1), 238–249. https://doi.org/10.1007/s11266-018-00087-z
Parker, M. A., Mook, L., Kao, C.-Y., & Murdock, A. (2020). Accountability and Relationship-Definition Among Food Banks Partnerships. Voluntas , 31 (5), 923–937. https://doi.org/10.1007/s11266-019-00150-3
Peci, A., Oquendo, M. I., & Mendonça, P. (2020). Collaboration, (Dis)trust and Control in Brazilian Manufactured Public/Non-profit Partnerships. Voluntas , 31 (2), 375–389. https://doi.org/10.1007/s11266-018-0027-6
Peralta, K. J., & Vaitkus, E. (2019). Constructing Action: An Analysis of the Roles of Third Sector Actors During the Implementation of the Dominican Republic’s Regularization Plan. Voluntas , 30 (6), 1319–1331. https://doi.org/10.1007/s11266-018-0003-1
Pettigrew, S., Jongenelis, M., Jackson, B., & Newton, R. U. (2019). “Charity Begins at Home”: Informal Caring Barriers to Formal Volunteering Among Older People. Voluntas , 30 (5), 921–931. https://doi.org/10.1007/s11266-018-0017-8
Pfeilstetter, R. (2020). Doing Good and Selling Goods. Voluntas , 31 (3), 511–520. https://doi.org/10.1007/s11266-020-00194-w
Pixová, M. (2018). The Empowering Potential of Reformist Urban Activism in Czech Cities. Voluntas , 29 (4), 670–682. https://doi.org/10.1007/s11266-018-0011-1
Pluciński, P. (2018). Forces of Altermodernization: Urban Social Movements and the New Urban Question in Contemporary Poland. Voluntas , 29 (4), 653–669. https://doi.org/10.1007/s11266-018-0007-x
Popplewell, R. (2018). Civil Society, Legitimacy and Political Space: Why Some Organisations are More Vulnerable to Restrictions than Others in Violent and Divided Contexts. Voluntas , 29 (2), 388–403. https://doi.org/10.1007/s11266-018-9949-2
Puljek-Shank, R. (2018). Civic Agency in Governance: The Role of Legitimacy with Citizens vs. Donors. Voluntas , 29 (4), 870–883. https://doi.org/10.1007/s11266-018-0020-0
Reijnders, M., Schalk, J., & Steen, T. (2018). Services Wanted? Understanding the Non-take-up of Social Support at the Local Level. Voluntas , 29 (6), 1360–1374. https://doi.org/10.1007/s11266-018-00060-w
Ruiz Sportmann, A. S., & Greenspan, I. (2019). Relational Interactions Between Immigrant and Native-Born Volunteers: Trust-Building and Integration or Suspicion and Conflict? Voluntas , 30 (5), 932–946. https://doi.org/10.1007/s11266-019-00108-5
Sandberg, B., Elliott, E., & Petchel, S. (2020). Investigating the Marketization of the Nonprofit Sector: A Comparative Case Study of Two Nonprofit Organizations. Voluntas , 31 (3), 494–510. https://doi.org/10.1007/s11266-019-00159-8
Schmid, H., & Almog-Bar, M. (2020). The Critical Role of the Initial Stages of Cross-Sector Partnerships and Their Implications for Partnerships’ Outcomes. Voluntas , 31 (2), 286–300. https://doi.org/10.1007/s11266-019-00137-0
Sepulveda, L., Lyon, F., & Vickers, I. (2020). Implementing Democratic Governance and Ownership: The Interplay of Structure and Culture in Public Service Social Enterprises. Voluntas , 31 (3), 627–641. https://doi.org/10.1007/s11266-020-00201-0
Shier, M. L., & Handy, F. (2020). Leadership in Nonprofits: Social Innovations and Blurring Boundaries. Voluntas , 31 (2), 333–344. https://doi.org/10.1007/s11266-018-00078-0
Spencer, S. B., & Skalaban, I. A. (2018). Organizational Culture in Civic Associations in Russia. Voluntas , 29 (5), 1080–1097. https://doi.org/10.1007/s11266-017-9925-2
Stewart, A. J., & Twumasi, A. (2020). Minding the Gap: An Exploratory Study Applying Theory to Nonprofit Board Management of Executive Transitions. Voluntas , 31 (6), 1268–1281. https://doi.org/10.1007/s11266-020-00244-3
Stougaard, M. S. (2020). Co-producing Public Welfare Services with Vulnerable Citizens: A Case Study of a Danish-Somali Women’s Association Co-producing Crime Prevention with the Local Authorities. Voluntas . https://doi.org/10.1007/s11266-020-00235-4
Strichman, N., Marshood, F., & Eytan, D. (2018). Exploring the Adaptive Capacities of Shared Jewish–Arab Organizations in Israel. Voluntas , 29 (5), 1055–1067. https://doi.org/10.1007/s11266-017-9904-7
Suter, P., & Gmür, M. (2018). Volunteer Engagement in Housing Co-operatives: Civil Society “en miniature”. Voluntas , 29 (4), 770–789. https://doi.org/10.1007/s11266-018-9959-0
Svensson, K., & Gallo, C. (2018). The Creation of an Unsolicited Organization: Victim Support Sweden. Voluntas , 29 (5), 1008–1018. https://doi.org/10.1007/s11266-018-9978-x
Tadesse, H. A., & Steen, T. (2019). Exploring the Impact of Political Context on State–Civil Society Relations: Actors’ Strategies in a Developmental State. Voluntas , 30 (6), 1256–1269. https://doi.org/10.1007/s11266-018-00077-1
Taylor, R., Torugsa, N. (Ann), & Arundel, A. (2020). Organizational Pathways for Social Innovation and Societal Impacts in Disability Nonprofits. Voluntas , 31 (5), 995–1012. https://doi.org/10.1007/s11266-019-00113-8
Uzcanga, C., & Oiarzabal, P. J. (2019). Associations of Migrants in Spain: An Enquiry into Their Digital Inclusion in the “Network Society” in the 2010s. Voluntas , 30 (5), 947–961. https://doi.org/10.1007/s11266-019-00162-z
van der Veer, L. (2020). Treacherous Elasticity, Callous Boundaries: Aspiring Volunteer Initiatives in the Field of Refugee Support in Rotterdam. Voluntas . https://doi.org/10.1007/s11266-020-00260-3
van Wessel, M., Naz, F., & Sahoo, S. (2021). Complementarities in CSO Collaborations: How Working with Diversity Produces Advantages. Voluntas , 32 (4), 717–730. https://doi.org/10.1007/s11266-020-00227-4
Vandepitte, E., Vandermoere, F., & Hustinx, L. (2019). Civil Anarchizing for the Common Good: Culturally Patterned Politics of Legitimacy in the Climate Justice Movement. Voluntas , 30 (2), 327–341. https://doi.org/10.1007/s11266-018-00073-5
Vuković, D. (2018). The Quest for Government Accountability and Rule of Law: Conflicting Strategies of State and Civil Society in Cambodia and Serbia. Voluntas , 29 (3), 590–602. https://doi.org/10.1007/s11266-018-9984-z
Waardenburg, M. (2021). Understanding the Microfoundations of Government–Civil Society Relations. Voluntas , 32 (3), 548–560. https://doi.org/10.1007/s11266-020-00221-w
Wells, R., & Anasti, T. (2020). Hybrid Models for Social Change: Legitimacy Among Community-Based Nonprofit Organizations. Voluntas , 31 (6), 1134–1147. https://doi.org/10.1007/s11266-019-00126-3
Williamson, A. K., Luke, B., & Furneaux, C. (2021). Ties That Bind: Public Foundations in Dyadic Partnerships. Voluntas , 32 (2), 234–246. https://doi.org/10.1007/s11266-020-00269-8
Yanay-Ventura, G. (2019). “Nothing About Us Without Us” in Volunteerism Too: Volunteering Among People with Disabilities. Voluntas , 30 (1), 147–163. https://doi.org/10.1007/s11266-018-0026-7
Yanay-Ventura, G., Issaq, L., & Sharabi, M. (2020). Civic Service and Social Class: The Case of Young Arab Women in Israel. Voluntas . https://doi.org/10.1007/s11266-020-00210-z
Zhang, Y., Bradtke, M., & Halvey, M. (2020). Anxiety and Ambivalence: NGO–Activist Partnership in China’s Environmental Protests, 2007–2016. Voluntas , 31 (4), 779–792. https://doi.org/10.1007/s11266-020-00237-2
Zihnioğlu, Ö. (2019). The Prospects of Civic Alliance: New Civic Activists Acting Together with Civil Society Organizations. Voluntas , 30 (2), 289–299. https://doi.org/10.1007/s11266-018-0032-9
Rights and permissions
Reprints and permissions
About this article
McMullin, C. Transcription and Qualitative Methods: Implications for Third Sector Research. Voluntas 34 , 140–153 (2023). https://doi.org/10.1007/s11266-021-00400-3
Download citation
Accepted : 18 August 2021
Published : 10 September 2021
Issue Date : February 2023
DOI : https://doi.org/10.1007/s11266-021-00400-3
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Qualitative methods
- Transcription
- Find a journal
- Publish with us
- Track your research

Qualitative Data Transcription: The Ultimate Guide
- March 27, 2024
SpeakWrite Blog
Discover the essentials of qualitative data transcription for businesses: unlock insights from interviews and customer feedback with accuracy and efficiency..

When it comes to mining qualitative data, the race to turn conversations into actionable insights is on. But let’s face it—transcribing all that information from interviews, focus groups, and customer calls can feel like trying to catch lightning in a bottle.
It’s slow, fraught with the risk of misinterpretation, and challenging to scale.
So, if you’ve ever found yourself bogged down by these transcription troubles, then you’re in the right place. Here’s what every business needs to know about qualitative data transcription.
What Is Qualitative Data Transcription?
Qualitative data transcription involves converting recorded audio or video of a subject into text format. This text can then be analyzed to identify patterns, themes, and insights that help you make crucial decisions about your business.
Qualitative data is different from quantitative data. Quantitative data is measured precisely with numbers, such as revenue in dollars or weight in pounds. Qualitative data , on the other hand, is observational, and includes open-ended responses rather than quantifiable ones.
Qualitative data transcription is often used by businesses to gather data from:
- Focus groups
- Customer support phone calls
- Product testing observations
- Court hearings
Uses Cases For Qualitative Data Transcription
Businesses thrive on deeply understanding their customers. Transcribing interviews , focus groups, and even customer review videos can unlock a treasure trove of consumer insights.
Market Research and Consumer Insights
Focus groups and consumer interviews are an important part of market research. The nuances, hesitations, emphatic endorsements—all these subtleties are captured in transcription, providing a rich, dimensional view of consumer sentiment.
Customer feedback can also inform the development of new features, products, or services that directly address customer desires, potentially opening up new markets or solidifying the business’s position in existing ones.
Customer Feedback Analysis
Transcribed customer feedback from support or sales calls can be used by businesses to use text analysis tools to identify recurring themes, sentiments, and emerging customer needs.
For instance, if multiple customers mention a specific defect with a product, this trend can prompt a targeted response, such as a product update or a change in customer service protocols.
Benefits of Qualitative Data Transcription Software & Services
Using qualitative data transcription tools and services is a game changer for many companies. Transcribing content in-house is exhausting, monotonous, and time-consuming, but a skilled transcription service can make the process quick and easy.
Professional data transcription services save time.
Professional transcriptionists can complete qualitative data transcription faster and more accurately than the average person. With the resources to transcribe at scale, professional transcription services quickly get the job done, giving you time to focus on other work.
Transcribing qualitative data improves accessibility.
Qualitative analysis usually requires several researchers, and transcribed data makes sharing content with your research team a breeze. Plus, anyone who can’t listen to audio can still effectively analyze content thanks to the process of transcription.
Professional data transcription is more accurate.
Professional transcribers can reduce the risk of errors in your written documents, improving the quality of your data. Accurate transcript information and grammar make all the difference when analyzing qualitative metrics, such as sentiment.
4 Types Of Qualitative Data Transcription
Here are a few different types of transcription.
Verbatim Transcription
With verbatim transcripts , everything is written down word-for-word. This includes:
- Nonverbal cues
- False starts
- Background noises
- Filler words like “um”
Considering experts suggest the majority of communication happens non-verbally , verbatim transcriptions are valuable to qualitative researchers.
This option is the most accurate to the content you send in, which is perfect if you want to ensure the speaker’s sentiment isn’t altered. But if you’re more interested in slicing through the fluff to get to the gist of things, verbatim transcription isn’t the best option.
| Hi, um, I recently bought your smart thermostat, and I, uh, can’t seem to get it connected to my Wi-Fi. It’s, like, really frustrating. |
| Oh, I’m really sorry to hear that. Uh, let’s see what we can do. Have you, um, have you tried resetting the device? |
| Yeah, I did, like, twice. It just, uh, keeps saying “Connection Failed” or something. |
Intelligent Transcription
Intelligent verbatim transcription includes all words from the file except unnecessary items, such as filler words and repetitions. This type of transcription makes the content more concise and easier to read for researchers who need to quickly skim the text.
| Hi, I recently bought your smart thermostat and can’t seem to get it connected to my Wi-Fi. It’s really frustrating. |
| I’m really sorry to hear that. Let’s see what we can do. Have you tried resetting the device? |
| Yes, I did twice. It just keeps saying “Connection Failed” or something. |
Edited Transcription
Edited transcription closely captures the speaker’s sentiment with generous tweaks to improve its quality and readability. It includes all the essential content from the audio file, but the words may be rearranged, condensed, or edited to make it more coherent and grammatically correct.
| Hi, I recently bought your smart thermostat and can’t get it connected to my Wi-Fi. It’s really frustrating. |
| I’m really sorry to hear that. Let’s see what we can do. Have you tried resetting the device? |
| Yes, I did twice. It keeps saying “Connection Failed.” |
Phonetic Transcription
Phonetic transcription includes phonetic symbols in the content to show how words are pronounced in the audio file. It can help clarify words with more than one pronunciation or a different pronunciation, depending on context.
How To Transcribe Data For Qualitative Research
Wondering how to transcribe data for qualitative analysis? Once you receive your transcription documents, it’s time to start coding, organizing, and managing your qualitative data . Coding transcripts in qualitative research involves four steps.
Step #1 – Initial Read-Through
Start by reading the entire transcript thoroughly to get a sense of the overall content and context. Coding is typically iterative, requiring multiple passes through the data to refine codes and themes.
Step #2 – Open Coding
Begin with open coding, where you label data segments with codes representing themes, ideas, or concepts as they emerge from the data. Take your time and perform as many read-throughs as you need.
Step #3 – Axial Coding
Next, it’s time to link codes to find relationships, categorizing them into broader themes or patterns.
Step #4 – Selective Coding
Selective coding involves focusing on key themes or concepts central to your research question, refining and integrating categories. Qualitative data analysis software can help manage and organize codes, making the process more efficient.
How To Choose The Right Qualitative Transcription Services
If you’ve been using manual transcription methods (i.e., transcribing content in-house), you’re no doubt wasting valuable time and resources. That’s where professional qualitative data transcription services can help.
To get started, you’ll need to pick the best business transcription service for your situation. Choose a transcription service to use by considering the following factors:
- Do they use human transcription instead of A.I. to help you perform an accurate qualitative analysis?
- Do they have experience with your industry?
- Can they accept your digital file type?
- What is the typical level of accuracy?
- Are their reviews positive?
- Do they offer customer support, if needed?
- Can the service include speaker names or time stamps, if needed?
- How quickly can they get it done?
The ideal service should be able to quickly transcribe the type of file you need using your preferred transcription type (verbatim, edited, etc.) Companies with human transcribers, excellent reviews, and fast turnaround times will give you the best results.
How To Use Qualitative Data Transcription Services
Once you choose a transcription company, it’s time to review their process. Typically, the process looks something like this:
- Step #1 – Choose the audio or video files you need transcribed.
- Step #2 – Upload the file to the transcription app. If you have any special requests for the process and results, note those requests in your instructions to the transcriptionist.
- Step #3 – Check your email—you should receive a written document with an accurately typed-out version of your recording. While you wait for the completed transcription, you’ll have extra time to focus on other work-related tasks (or kick back and take a well-deserved break!)
The Best Qualitative Data Transcription Services
If you’re looking for fast and accurate qualitative data transcription, SpeakWrite has you covered. Their human transcriptionists are known for being extremely thorough and detail-oriented.
Unlike A.I. transcription companies that guarantee only up to 85% accuracy, SpeakWrite offers 99% to 100% accuracy verbatim transcriptions. Its transcriptionists are available 24 hours a day and offer a standard turnaround time of about 3 hours.
You can submit a variety of document types that will remain confidential thanks to the site’s multi-level security with authorization. All the typists are also fully vetted with strict background checks for your safety.
Instead of paying monthly fees like you would with other companies, SpeakWrite only charges you for completed work at a per-word rate: no contracts, no surprises, no hidden charges.
How To Get Started With Qualitative Data Transcription Services
Starting qualitative data transcription services with SpeakWrite is easy. You can get started in three simple steps:
- Step #1 – Create an audio recording.
- Step #2 – Submit your audio file through the app, by email, or by using the toll-free dictation line anytime.
- Step #3 – Receive a completed document in about three hours.
Qualitative Data Analysis Transcription: Frequently Asked Questions:
How do you transcribe data for qualitative research.
For most businesses, it makes sense to hire a qualitative data transcription company to do the work for you. All you need to do is decide whether to transcribe verbatim (including every word and sound) or opt for an edited transcript that omits filler words and irrelevant content.
If you are transcribing your own content, consider using transcription software or services to speed up the process. These tools can automatically transcribe audio to text, though human verification and editing are often necessary to ensure accuracy.
For high accuracy, especially in cases of complex terminology or poor audio quality, manual transcription may be preferred.
What is an example of verbatim transcription in qualitative research?
An example of verbatim transcription would capture every word, pause, laugh, and non-verbal sound in the recording.
For instance, if a participant says, “Well, uh, I think—you know—it’s really quite hard to say. [laughs] I mean, everything is fine, I suppose,” a verbatim transcript would include all of those hesitations, fillers, and laughter exactly as they occur.
Based on how the speaker chose their words in the quote above, everything is not, in fact, fine. For qualitative researchers, all the filler words, stammering, and nonverbal cues help them “read between the lines.”
What is the most common form of transcription in qualitative interviews?
The most common form of transcription in qualitative interviews is intelligent verbatim transcription. This approach includes meaningful speech content while omitting filler words, false starts, and other non-verbal or irrelevant elements (e.g., “uh,” “um,” “you know”).
It strikes a balance between capturing the essence of what was said and readability, making it practical for most qualitative research purposes.
How is confidentiality maintained during the transcription process?
Ensuring confidentiality in the transcription process involves several key practices. Firstly, choosing a reputable transcription service that adheres to strict confidentiality agreements is crucial. These services often implement secure file transfer protocols to protect data during transmission and storage.
When transcribing in-house, it’s essential to educate all team members on confidentiality protocols. This includes anonymizing participant information, using secure networks for file transfers, and storing data on encrypted devices.
Additionally, confidentiality can be maintained by:
- Assigning unique identifiers to participants instead of using personal information.
- Securing informed consent forms that detail how data will be used and protected.
- Implementing strict access controls to ensure that only authorized personnel can view sensitive data.
Get Started With Qualitative Data Transcription Today
Businesses and researchers have trusted SpeakWrite with their qualitative data transcription since 1997. Since the very beginning, SpeakWrite has transcribed files using 100% human transcriptionists.
That means ready-to-use professional transcriptions already formatted and edited for accuracy, 100% of the time. Place an order today to take advantage of Speakwrite’s trusted qualitative data transcription services.
Explore FAQs
Discover blogs, get support.


Data Transcription in Qualitative Research - Everything You Need To Know
Speak content team.
- October 22, 2021
Working with language data? Save 80%+ of your time and costs.
Join 150,000+ individuals and teams who rely on speak ai to capture and analyze unstructured language data for valuable insights. streamline your workflows, unlock new revenue streams and keep doing what you love..
Get a 7-day fully-featured trial!

Data Transcription in Qualitative Research: Everything You Need to Know
Learn about why you need data transcription in your qualitative research process and how to pick the right transcription services for you..
If your work involves data transcription in qualitative research, you might be overwhelmed by the amount of language data you record and transcribe on a daily basis.
Historically, transcription has always been the work of professional transcribers because of the high accuracy standards required for research validation.
These days, data transcription is increasingly going digital, with AI transcription looking to disrupt the space . This is primarily due to their lower costs, more efficient transcription workflow, and assistive features for professional transcribers.
Table of Contents
What is data transcription in qualitative research.
Data transcription refers to converting speech into written text for documentation or analysis purposes. Data transcription is also the first and most important step in a qualitative research project .
Before analyzing any recorded audio, you must first convert them into text. Once these transcripts are available, you can go through them multiple times and pick up on valuable unanticipated patterns.
Examples of dialogues that require transcription include:
- Focus group studies
- Patient consultations
- Court hearings
Since qualitative research is focused on exploring subjective characteristics and cannot be measured by numbers, a clean language data set is critical. Without a clean data set, your analysis could be incomplete or completely inaccurate.
Types of data transcription
There are two main types of data transcription - verbatim and intelligent transcription.
Verbatim/true verbatim transcription
What is verbatim transcription.
The first type of transcription is verbatim transcription, which means recording every part of the dialogue or sound on the audio file. This includes grammatical errors, pauses, and even non-verbal communication.
Verbatim transcription is often the preferred method for subjective qualitative research due to its 100% accuracy in staying true to the speaker’s intention. Rather than recording for readability, it focused on capturing how the person spoke.
Here’s an example:
Interviewer: So, you mentioned that you had some (coughs) experiences with this product. Could you please elaborate more on that?
Participant: Yeah, we bought the product and the entire family [laughs] loved it.
By capturing mannerisms in addition to the words spoken, readers can gain a more holistic understanding of the interview environment and rapport between the participants.
When is verbatim transcription necessary?
Verbatim transcription is necessary when your research’s goal is to capture the participant’s subjective feelings. By recording non-verbal cues and the research environment, you can interpret the speaker’s intentions more comprehensively.
Situations where you should use verbatim data transcription in qualitative research include:
- Focus groups or group discussions
- Patient diagnosis
According to various studies, up to 70-93% of all communication is nonverbal . While this “rule” is difficult to prove with empirical results, the intent is clear -- analyzing communication is more than just looking at the words used . You must also consider the non-verbal cues and tonality to analyze the speaker’s sentiments accurately.
One way to record verbatim transcription is through the help of professional transcribers, such as the human transcription services provided by Speak Ai .
Once you have a 99%+ accurate transcript, you can extract actionable insights using our software with more confidence, knowing that your language data set is as precise as possible
Intelligent verbatim/edited transcription
What is intelligent verbatim transcription.
Intelligent verbatim (also called clean or edit transcription) is transcription that omits fillers, repeating words and grammatical errors. The main focus of intelligent verbatim is to optimize the transcript for readability and clarity.
Intelligent verbatim transcription captures what the speaker is communicating rather than the how.
Here is an example:
Interviewer: So, you mentioned that you had some good experiences with this product. Could you please elaborate more on that?
Participant: Yeah, we bought the product and our entire family loved it!
As you can see, intelligent verbatim transcription improves the conciseness and clarity of the text. However, it can also unintentionally alter the original meaning of the sentences.
A professional transcriber usually does intelligent verbatim transcription. On the other hand, you can also use automated data transcription tools if your priority is to save time or just to get a mostly accurate first draft.
Automated data transcription is much more affordable for qualitative research. However, you may require some light editing to fix errors the speech-to-text AI makes.
When are edited transcripts necessary?
You should use intelligent verbatim transcription if you need transcripts that are easy to read or skim through. These transcripts may not record the non-verbal cues or sentiments of the speaker as well, but are perfect for capturing the dialogue content.
Moreover, you can easily share intelligent verbatim transcription because of its conciseness. Since the recordings have been optimized for reading, intelligent data transcription also has more practical applications and can be used for more general purposes.
Situations where intelligent verbatim could be more suitable include:
Team meetings
Publicly available transcripts
Patient records
General business purposes
How to conduct data transcription for qualitative analysis?
There are two main ways to transcribe data for qualitative research purposes.
Professional human transcription is the best way to ensure the most accurate transcript possible -- though it is more expensive and takes a lot more time to complete. This is usually reserved for important files and research jobs where data accuracy comes first.
For individuals or businesses with less stringent requirements for the accuracy of their data set, it is possible to supplement or entirely replace human transcription with automated transcription software.
Professional human transcription services for qualitative research
While automated speech recognition technologies are steadily improving, manual transcription is still superior for delivering accurate transcripts.
Anyone can transcribe an audio recording. However, it takes the average person anywhere from 4 - 5 hours to transcribe one hour of audio accurately. In comparison, a trained transcriptionist can complete the same job in as little as 2 hours.
If you work in research where you are appraised more on your research output than time put in, hiring a professional transcriptionist is probably a better use of your time. This way, you can focus your efforts on the work that matters -- analyzing the transcripts for key moments that matter and converting them into actionable insights.
Unlike automated transcriptions of qualitative data, human transcription is usually 99-100% accurate. Labor costs can also vary depending on where the professional is based, costing anywhere from $0.50 up to $2.00 per audio minute .
Depending on the size of the workload and urgency of the job, Speak Ai ’s expert team of transcribers can deliver your transcript in as little as 48 hours. You can also utilize our software’s advanced analysis tools, such as keyword extraction and sentiment analysis, to identify key moments in your text as part of your subscription.
Automated data transcription for qualitative research
Automated transcription services may not guarantee the same level of accuracy as professional transcribers, but they are a great way to scale your transcription efforts. Additionally, if the file audio quality is good, it is possible to get up to 95% accuracy with AI-powered transcripts - with only a small amount of transcript cleanup required.
There are various automated transcription tools available in the market. However, with Speak Ai, you get an all-in-one transcription software that significantly enhances your workflow. You can get automated transcriptions starting as low as $0.15 per audio minute , a 1:1 turnaround time, and an integrated option to order professional help within the platform to clean up your transcripts.
Not to mention a whole suite of machine-powered analyses to supplement your media transcripts as well as researcher and developer-friendly options, including JSON and CSV.
Automated transcription services are a great starting point to build an accessible media database. Once you have a fully transcribed media library, you can decide which of your files are important enough to get professionally transcribed if you need cleaner data sets.
How to organize data transcription in qualitative research?
As you collect more data over time, your transcription management system can turn into a disorganized mess when using outdated workflows. Using an integrated transcription solution can save you many headaches down the line by letting you organize, transcribe, analyze, store and access your files by project, all in one place.
Here are some tips on how you can organize transcribed data neatly and extract key information more efficiently.
Keyword mapping
Keyword mapping refers to identifying and organizing prevalent keywords in a presentable manner. You can manage these keywords in an excel sheet or with an automatically generated word cloud.

Keyword mapping is an effective method of identifying patterns both in your research and your competitors’ content. This insight on trending topics, keywords, and other relevant named entities will allow you to make conclusions with a more holistic overview.
Try Our AI Word Cloud Generator
Word clouds are a great way to highlight the most important words, topics and phrases in a text passage based on frequency and relevance. Generate word clouds from your text data to create an easily understood visual breakdown for deeper analysis. Try our free word cloud generator today to automatically visualize insights from your data.
Utilize data management systems
Extensive qualitative research entails coordination between different teams and departments across geographical regions. Simply storing your transcriptions in a cloud drive or email chains may work for a while but will clutter up quickly.
Robust data management systems such as Tableau allow you to share qualitative data results with anyone in your company, anywhere, at any time.
Using Tableau in conjunction with a transcription and QDA tool like Speak Ai lets you create a centralized, searchable media library to supplement your research process. By building this data management system, you can break down silos, enhance collaboration, and better validate your research findings in a streamlined manner.
Establish transcription procedures and requirements
Different transcribers may have different styles of converting audio speech into text. While this is not an issue in small teams, clashing transcription styles will cause confusion when more transcribers get involved.
Before you begin data transcription for qualitative research or any other purposes, ensure that you address:
- The type of transcription (verbatim or intelligent verbatim)
- Formatting consistencies
- Quality control procedures
- Translations
- Any other specific transcription instructions
- Building a custom vocabulary list for industry specific terms
- Ethical and legal considerations
Clear transcription and quality control procedures may be a hassle to establish at first. However, good data and work organization always pays dividends in the long run and optimizes workflows.
Who can benefit from data transcription services?
If you handle large amounts of video and audio recordings and need to convert them into text, you will need a data transcription service.
Market researchers: Market researchers can use verbatim data transcription (word-for-word) for primary data sources such as group interviews. They can then access the transcripts as needed in their research process. Having an easily searchable, centralized library of all recorded interviews reduces administrative time and ensures better research results.
Medical: Medical professionals can benefit from data transcription in many ways. For example, patient records, surgery notes, and medical procedures can all be transcribed to ensure that no critical diagnostic information is missed.
Business: You can use automated data transcription services to make taking notes a breeze. Common occurrences of data transcription in business settings include recording stakeholder meetings, consultations, and client meetings for you to analyze after.
Legal: Legal transcripts are commonly used to record all parts of court proceedings, such as arguments, defense, and judge’s decisions.
Academic: Students and educators alike can use data transcription services to record lectures, consultations, presentations, dissertations, and other research work.
tl;dr - Key Takeaways
Data transcription in qualitative research is the first and most crucial step of the research process (and often the most expensive).
The best way to ensure a 99% accurate language data set for qualitative research studies is still through professional transcription services . However, automated transcription services are becoming more commonplace to make this process more efficient.
Medical professionals, market researchers, enterprises, lawyers, students, and educators can all benefit from an effective data transcription service when looking to optimize their research workflows.
If you are interested in finding out how to utilize data transcription in your qualitative research, sign up for Speak Ai’s 7-day trial today to get transcription management, qualitative data analysis, and shareable media databases all in one place.
Join a growing number of enterprises , researchers , and marketers using Speak Ai to make better decisions, faster.
Get a 7-day fully-featured trial.
Save 99% of your time and costs!
Use Speak's powerful AI to transcribe, analyze, automate and produce incredible insights for you and your team.
Data Transcription in Qualitative Research: All You Need to Know
Qualitative research transcription is a critical step in the research process where recorded interview audio is converted into text. According to a study by Oxford Academic, data transcription is the first step in analyzing data for qualitative research . After all, with it, you can better examine what was said in the interviews you conducted. That way, you can gather more relevant data for your study. It also makes said data easier to analyze and share with your team, allowing data analysis to proceed much faster.
Additionally, with such transcriptions on hand, patterns can be easier to find, and researchers can use said patterns and data to create a narrative for their study. If you plan to conduct data transcription, it’s always a good idea to learn more about it beforehand. Here’s everything you should know about data transcription in qualitative research.
The Two Types of Data Transcription
Verbatim transcription.
One of the types of data transcription that researchers can consider is verbatim transcription. When doing verbatim transcriptions, researchers transcribe every part of the interview audio. Apart from the dialogue, these include spoken grammatical errors, pauses, and even sounds such as laughing or coughing.
Often, verbatim transcription is done if the qualitative research is subjective. This kind of transcription can convey the speaker’s intention with incredible accuracy. By focusing more on how the speaker talks, readers are afforded a better understanding of the discussion. It also allows for a further grasp of the participants’ rapport with one another.
If you're opting for subjective research, you must capture the interviewee’s personal feelings. As such, be sure to have your human transcriptionists use verbatim transcriptions to record the dialogue and the non-verbal cues in the audio. That way, you can use the tone to analyze the speaker’s intentions and sentiments comprehensively.
Intelligent Verbatim Transcription
Intelligent verbatim transcriptions are also known as clean or edited transcriptions. As its name suggests, this is a type of transcription that removes fillers and grammatical errors with the goal of making the transcript more readable and concise. Intelligent transcriptions focus more on the information provided rather than the interviewee's subjective feelings.
Intelligent verbatim transcriptions are also best used if you need the transcript to be easy to read or skim through. This is because, compared to verbatim transcriptions, it doesn’t need to capture the interviewee’s non-verbal cues. As a result, the final transcription is more optimized for reading and can be used for general purposes.
This type of data transcription is also best used for objective qualitative or quantitative research. By opting for intelligent verbatim transcriptions, the cleaned and edited transcripts will help researchers analyze the interview more accurately for their study.
However, keep in mind that this type of transcription might alter the original meaning of what the speaker is trying to convey. As such, whether you transcribe the audio yourself or enlist the help of academic transcription services , it’s best to carefully consider whether intelligent transcriptions are the best for your research.
How to Organize Data Transcription in Qualitative Research
The more you transcribe the interviews you conducted, the more data you can gather for your research. However, an abundance of data can result in a disorganization of the materials you need. Thankfully, there are ways you can organize the data you compile for your qualitative research.
Keyword Mapping
Keyword mapping is one method of data organization that researchers can consider. Usually, this refers to the identification of keywords that are prevalent in your transcript. Once the keywords are identified, you can begin to organize them in a spreadsheet. On the other hand, you can also take advantage of transcription apps with word cloud capabilities to identify or organize your keywords.
Take Advantage of Data Management Systems
Another method of data transcription organization you can consider is using data management systems. Remember, when conducting qualitative research, coordination and collaboration within your team and the relevant departments are essential. However, these teams and departments are sometimes located elsewhere in the country or even the world. As such, researchers must take advantage of data management systems to organize their data transcripts.
By using data management systems, your qualitative research transcription can be stored in a centralized media library that anyone in the team can access. As a result, research collaboration is improved, and the research can be validated effectively.
Follow Transcription Procedures and Requirements
Finally, when organizing your data transcription, it’s highly recommended that transcription procedures and requirements be followed. This is especially important if the team isn't availing of a human transcription service . If the team is transcribing the audio themselves, differing transcription styles can cause confusion. As such, before you begin transcribing your audio files, be sure to discuss the following with your team members:
- Type of transcription
- Consistent formatting
- Quality control procedures
- Translations (if needed)
- Specific transcription instructions
- Custom vocabulary list for industry-specific terms
By considering the above-mentioned items, the team can have more accurate transcripts for the study. The precise transcription will allow the data you gather to be more organized. In addition, your team's workflow will also be fully optimized, allowing for easier collaboration.
Transcriptions are a vital part of gathering data for academic research endeavors. However, it’s also a time-consuming process. As such, turn to TranscriptionWing for your academic transcription needs.
We have an expert team of transcriptionists to cater to your needs. Additionally, we also offer machine cleaning services for AI-generated transcripts. With this service, the team will remove spelling, grammatical, and punctuation errors, increasing your transcript’s readability and coherence.
With TranscriptionWing, clean qualitative research transcription is well within reach. Learn more about our services by contacting us today !
Related Posts
Free recording service.
We use cookies to ensure that we give you the best experience on our website. If you continue to use this site we will assume that you are happy with it. Privacy policy
Qualitative Research Best Practices: The Researcher’s Guide to Accurate and Insightful Transcription
Qualitative research is an essential methodology in the social sciences , allowing researchers to capture the nuances of human experiences, behaviors, and interactions. Unlike quantitative research, which seeks to quantify data, qualitative research focuses on understanding the significance and meanings people attribute to their social reality. Accurate transcription is a critical component in qualitative research as it ensures that the spoken word is faithfully converted into written text, preserving the subtleties and context of the conversation.
This attention to detail provides a solid foundation for analysis , ensuring that interpretations and conclusions drawn are rooted in what participants have actually communicated . The purpose of this guide is to provide a thorough overview of the best practices for transcription in qualitative research and to highlight the impact of accurate transcription on the integrity of research findings . By understanding and employing rigorous transcription techniques, researchers can maintain the fidelity of their data, making certain that analyses and subsequent knowledge generated are both credible and reliable.
Table of Contents
1- Understanding Transcription in Qualitative Research 2- Best Practices for Accurate Transcription 3- Cultivating Insightful Transcription 4- Ethical Considerations in Transcription 5- Future Trends and Innovations in Transcription 6- To Wrap Up
Understanding Transcription in Qualitative Research
The process of transcription in qualitative research is a meticulous endeavor that demands a deep understanding of the communicative intricacies captured during data collection. It serves as more than just a methodological step; it is a critical activity that shapes the direction of data analysis. At its core, transcription is the act of translating oral communication into written form, creating a tangible representation of conversations, interviews, or focus groups that researchers can methodically examine.
This translation plays a pivotal role in qualitative data analysis as it converts ephemeral spoken words into a format that allows for detailed coding, thematic exploration, and the unravelling of conceptual understandings. There’s a spectrum of transcription methods available to researchers, ranging from verbatim, which captures every utterance and sound, to intelligent verbatim, which streamlines the speech by omitting non-verbal communication and irrelevant information, thus providing a clear, concise record. Choosing the appropriate transcription method hinges on the research objectives and the level of detail necessary for analysis.

However, transcription is not without its challenges and common pitfalls. Researchers often grapple with issues such as distinguishing between relevant and irrelevant data , maintaining participant confidentiality, and contending with audio quality, accented speech, or overlapping dialogue. These factors can significantly impact the accuracy of the transcription and, by extension, the quality of the research findings . As such, being cognizant of these challenges, and applying best practices throughout the transcription process, is vital to uphold the authenticity and integrity of the data and ensure robust qualitative analysis.
Best Practices for Accurate Transcription
To begin with best practices for accurate transcription in qualitative research, preparation plays a pivotal role. Organizing data and materials beforehand ensures a smooth and efficient transcription process. Researchers must gather all the audio recordings, notes, and any additional materials that can provide context to the dialogue. Once organized, the next logical step is to select the appropriate transcription software or tools that meet the specific needs of the project, considering factors such as compatibility with audio formats, ease of use, and the ability to handle multiple speakers.
As the transcription process commences, active listening skills are paramount, as they enable the transcriptionist to discern subtle nuances in tone, emotion, and emphasis which might be crucial for the research analysis. Employing strategies for handling difficult audio quality or accents such as adjusting playback speed, using quality headphones, or segmenting difficult audio passages for closer attention, can greatly enhance the accuracy of the transcription . Researchers should also remain vigilant to maintain neutrality and avoid inadvertently introducing biases into the transcription by staying as true to the source as possible.
Ensuring transcription accuracy is an ongoing process, requiring the transcript to be double-checked and verified against the original audio to rectify any discrepancies. Dealing with ambiguous or unclear speech is a common challenge, where the context can often provide clues for interpretation; however, researchers should note any uncertainties within the transcript for clarity . Quality control measures, including peer reviews of transcripts or the use of software with accuracy-checking features, should be implemented to uphold the expected standard. This comprehensive approach to preparing for and conducting the transcription process is critical for preserving the integrity of qualitative research findings.

Cultivating Insightful Transcription
Cultivating insightful transcription within the realm of qualitative research is an intricate dance between rigor and creativity . It requires a deep understanding of the research context to breathe life into the transcribed text, allowing themes, patterns, and nuances to emerge with clarity. This synergy between transcription and analysis is paramount; as researchers immerse themselves within the data, they become adept at uncovering underlying meanings and insights that might otherwise remain concealed within the spoken word.
By integrating transcription with qualitative analysis techniques, such as coding and thematic analysis , the transcription process evolves into an interpretive act that sets the stage for rich data interpretation. One of the foremost goals is to transform the static text into a compelling narrative that faithfully represents participants’ experiences and perspectives, thus providing a voice to their stories.
In this way, transcription extends beyond a mere administrative task and becomes an essential tool in the qualitative researcher’s kit, allowing them to leverage the full depth and breadth of the data in the storytelling process. The cultivated transcripts are not just records of spoken words; they serve as a canvas upon which the researcher can paint a picture that brings research findings to life, thereby engaging with broader audiences and contributing to the collective understanding of the complex social world they are studying.
Ethical Considerations in Transcription
In the sphere of transcription within qualitative research , ethical considerations are paramount, as they directly affect the integrity of the process and the wellbeing of participants. Protecting participants’ confidentiality and privacy is a fundamental duty; it demands stringent measures, such as anonymizing transcripts, to shield identities and personal details from unwarranted disclosure. This task becomes even more crucial when dealing with sensitive or potentially harmful content, which must be handled with the utmost care and sensitivity —if necessary, by employing content warnings or sensitive data handling protocols to minimize harm or distress to participants or readers.
Alongside these protective practices stands the cornerstone of ethical research—obtaining informed consent for transcription and subsequent data use. Researchers must ensure that participants are fully aware of how their words will be used, their ability to remain anonymous, and their right to withdraw at any point without repercussion. Furthermore, the ethical landscape of transcription is not complete without addressing issues of power dynamics and researcher reflexivity. Power dynamics, especially between the researcher and the participants, can inadvertently shape the data collected and thus, researchers must continuously reflect on their positionality , potential biases, and influence on the data to ensure that the transcriptions present an unbiased and accurate reflection of participants’ voices.
Reflexivity involves a commitment to self-awareness and adaptability , acknowledging and adjusting the research approach in response to the evolving context. Together, these ethical tenets form a framework that guides researchers in conducting transcription with a balance of scientific rigor and humanistic concern, ultimately striving to uphold the dignity and respect of all involved parties throughout the research endeavor.

Future Trends and Innovations in Transcription
As we gaze into the horizon of qualitative research, emerging technologies in transcription beckon with promises of unprecedented efficiency and enhanced accuracy. Artificial Intelligence (AI) -driven transcription services are at the forefront of this revolution, employing sophisticated algorithms and machine learning to seamlessly convert speech to text .
Advancements in speech recognition and natural language processing have taken leaps forward, enabling these systems to parse complex language, discern nuances, and even distinguish between different speakers with greater precision than ever before. The potential impact on qualitative research methodologies and practices is profound ; with AI automation, researchers can allocate more time to the substantive analysis of content rather than the meticulous labor of transcription.
These technological strides also mean that transcripts can be generated swiftly, allowing for near real-time analysis that may uncover insights more readily. As AI continues to evolve , it holds the potential to further refine the interpretive processes intrinsic to qualitative analysis, potentially offering researchers not just transcribed text but preliminary thematic categorizations and sentiment analyses.
Such innovations could enable a more dynamic and iterative approach to data collection and analysis, blurring the lines between transcription and deep analysis and thereby enriching the storytelling capacity of qualitative research. The future of transcription technology thus offers a tantalizing glimpse of a more streamlined, powerful, and nuanced tool in the qualitative researcher’s arsenal, capable of capturing the human voice with an artistry and depth that edges ever closer to the richness of face-to-face conversation.

In conclusion, as we have traversed the nuances of qualitative research transcription , it’s evident that this practice is far from a mechanical task—it is an art that requires diligent attention to detail, a steadfast commitment to ethical standards, and an open-minded engagement with emerging technologies. From the initial capture of auditory data to the meticulous transformation of spoken words into written form, transcription serves as the backbone of qualitative analysis—structuring narratives, revealing participant experiences, and nurturing insights.
The integrity of this process hinges on strict adherence to best practices, including respecting participant confidentiality, leveraging accurate and unbiased transcription methods, and incorporating reflexivity. Indeed, the accuracy of transcription is not simply a measure of fidelity to audio but is instrumental in drawing out the rich tapestry of meaning that qualitative research seeks to understand.
As researchers, there is a continuous imperative to refine transcription skills, to adapt to the evolving landscape of technology, and to balance the precision of science with the empathy of human inquiry. Emboldening this journey is the promise of AI and speech recognition advancements which aim to amplify our analytical capabilities, not to replace the human element, but to support it. It suffices to say; the mastery of transcription is a testament to the researcher’s pursuit of clarity and commitment to elevating the human voice, a pursuit that, when approached with dedication and foresight, lends itself to the grand mosaic of qualitative scholarship and the collective quest for knowledge.
Interesting topics
- How to add subtitles to a video? Fast & Easy
- Subtitles, Closed Captions, and SDH Subtitles: How are they different?
- Why captions are important? 8 good reasons
- What is an SRT file, how to create it and use it in a video?
- Everything You Need for Your Subtitle Translation
- Top 10 Closed Captioning and Subtitling Services 2023
- The Best Font for Subtitles : our top 8 picks!
- Davinci Resolve
- Adobe After Effects
- Final Cut Pro X
- Adobe Premiere Rush
- Canvas Network
- What is Transcription
- Interview Transcription
- Transcription guidelines
- Audio transcription using Google Docs
- MP3 to Text
- How to transcribe YouTube Videos
- Verbatim vs Edited Transcription
- Legal Transcriptions
- Transcription for students
- Transcribe a Google hangouts meeting
- Best Transcription Services
- Best Transcription Softwares
- Save time research interview transcription
- The best apps to record a phone call
- Improve audio quality with Adobe Audition
- 10 best research tools every scholar should use
- 7 Tips for Transcription in Field Research
- Qualitative and Quantitative research
- Spotify Podcast Guideline
- Podcast Transcription
- How to improve your podcasting skills
- Convert podcasts into transcripts
- Transcription for Lawyers: What is it and why do you need it?
- How transcription can help solve legal challenges
- The Best Transcription Tools for Lawyers and Law Firms
Transcribing interviews for qualitative research

Transcribing interviews is an important step in qualitative research, as it forms the backbone of data analysis and interpretation. In other words we can say that it acts as a vital link between those unfiltered conversations and insightful data acquired from them. But why is accurate transcription so crucial in qualitative studies?
The fundamentals of qualitative research itself provide the first justification. The depth with which linguistic expressions and emotions are communicated during interviews is crucial for this kind of research. Accurate transcription ensures that these non-verbal cues are also added for more clarity.
Transcribing interviews qualitative research is essential to ensuring the correctness of findings because it enables researchers to fully capture the range of participant replies and perspectives. Moving forward in this article we have compiled a comprehensive guide to help you get a more clear perspective on how to transcribe interviews for qualitative research.
What Is qualitative research?
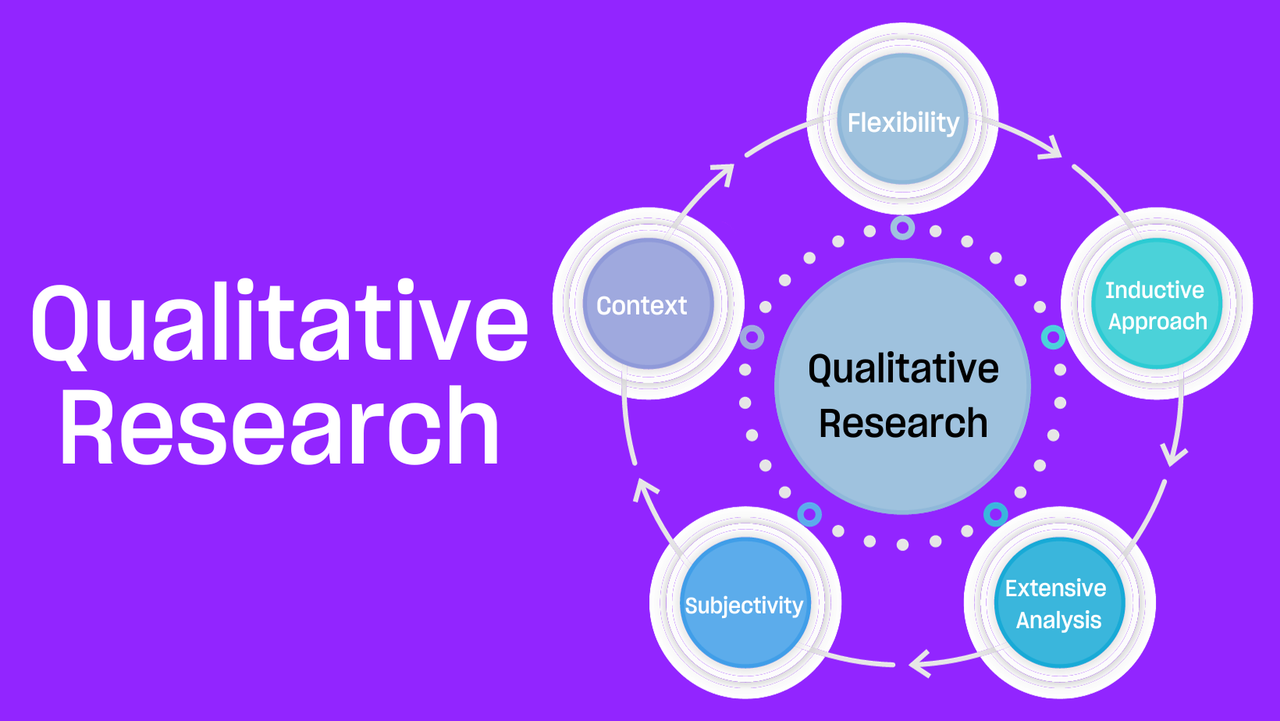
Qualitative research is one of the most commonly used research methods in the field of academia. Instead of concentrating just on the what, where, and when of decision-making, it explores the why and how by focusing on the human aspects of a specific issue or situation. It aims to comprehend people's experiences, actions, feelings, and the interpretations they place on objects.
Getting a much deeper insight into people's attitudes, actions, value systems, concerns, motives and goals is the main aim of qualitative research. It is employed to acquire a deeper comprehension of intricate occurrences that are challenging to put into numerical form.
The main characteristics of qualitative research are:
- Focus on context: It explores the context in which behaviours and events take place.
- Subjectivity: It recognises the subjective nature of the study and frequently captures the perspectives of the participants.
- Extensive analysis: This entails a thorough examination of a limited number of case studies or circumstances.
- Inductive approach: The inductive approach often begins with observations and builds theories from them.
- Flexibility in design: As the study goes on, the research question format may change. Here it is not necessary to follow the predetermined context.
Researchers use qualitative interview as their main method of data collection for this research since it allows them to interact with the subject first hand and focus on the non-verbal cues along with the information they are sharing.
Looking for support in transcribing your qualitative research interviews? Good Tape offers transcription services that can help you better understand your interviews. We're here to help make your transcription process more manageable and efficient. Explore how Good Tape can assist you in your research endeavors .
Qualitative vs quantitative interviews

Qualitative and quantitative interviews are different research approaches, each with a unique strategy for collecting and interpreting data. Quantitative interviews seek to measure human behaviour and experiences in a form that can be statistically examined, whereas qualitative interviews concentrate on investigating and comprehending the depth and complexity of human behaviour and experiences.
While both are extensively used in the field of research, it is important to understand where either of the two should be used. Below is a comparative table of both against which you can determine which of the two would work best in your scenario.
Aspect | Qualitative Interview | Quantitative Interview |
Purpose | To understand people's experiences, ideas, and feelings on a deeper level. | To quantify information and generalise findings to a wider audience. |
Nature of data collection | Textual, descriptive | Numerical, statistics |
Data collection method | Open-ended questions and unstructured interviews | Close-ended questions and structured interviews |
Sample size | Smaller since more focus is on depth of information | Larger since more focus is on statistics and generalisation |
Approach | Exploratory | Conclusive and factual |
Outcome | Detailed understanding of a particular problem or area of interest | Quantifiable insights |
Flexibility | Increased flexibility as questions are adapted according to the situation | Decreased flexibility as questions are predetermined and close-ended |
This table presents a clear contrast between qualitative and quantitative interviews, highlighting the differences in their technique, strategy, and study conclusions. The choice between both majorly depends on the research question at hand and the nature of the topic being studied.
How to transcribe an interview for qualitative research

For qualitative research, transcription of interviews is a painstaking procedure that needs time and close attention to detail. It requires turning spoken words from your recorded audio or video into text.
In qualitative research, this transcribing procedure is essential to data processing. Here's a step-by-step tutorial on effectively transcribing interviews, along with a few tips to make the process as easy as it can be.
Record clear audio of the interview
Select a peaceful, quiet workstation for your interviews to reduce distractions and improve focus. It is important to have a well-positioned microphone and high-quality headphones if you want to record even the minute details of speech without picking up excessive background noise.
If there are any unpleasant noises in your audio, services like Good Tape can be quite helpful. They are made to carefully pick up on all spoken and nonverbal cues, even in busy settings, and automatically transcribe all your work for you, so you won't miss any important information.
Work around your transcription
Precise transcription is essential for detailed analysis, accurately recording each word and nonverbal cue. This comprehensive approach allows for a deeper understanding of both the verbal as well as non-verbal cues in communication.
Similarly, intelligent verbatim concentrates on streamlining the text by eliminating unnecessary words and sounds to focus on the primary concepts, resulting in a transcript that is more focused and structured. Revised transcriptions enhance the material by improving clarity and fixing grammar, guaranteeing that the final transcript is accurate, comprehensible, and cohesive.
Audio transcription services such as Good Tape make accurate transcription easy with a shorter turnaround time.
Finalise the transcript
For easy navigation and the identification of important points or sensitive parts within the text, transcript formatting consistency is essential. Consistent formatting facilitates reading and improves the transcript's overall usefulness.
A further crucial stage is anonymisation, which anonymises any confidential or private data to comply with legal regulations. This also gives the interviewees peace of mind knowing that the information they provide will not be used illegally. To ensure that the transcript is correct, well-written, and presented professionally, one last review is necessary to spot any spelling, grammatical, or flow errors.
Some useful tips
Manual transcription can take a lot of time, therefore patience is essential. However, if you wish to have accurate transcripts in less time, using services such as Good Tape can cut down on the amount of time required.
It's also very important to make sure that your transcribed documents are safe. Maintaining regular backups is essential to avoiding data loss. Using services that automatically store and back up your transcribed audio might be a sensible choice if you find it difficult to remember to do backups, since they provide efficiency and peace of mind.
Why accurate transcription matters in qualitative research
Precise transcription is essential to qualitative research because it supports the accuracy and essence of the whole research process. It is the first stage of data analysis and has a direct impact on the findings and recommendations of the study. There are several reasons why accurate transcribing is important and advantageous.
Impact on data analysis
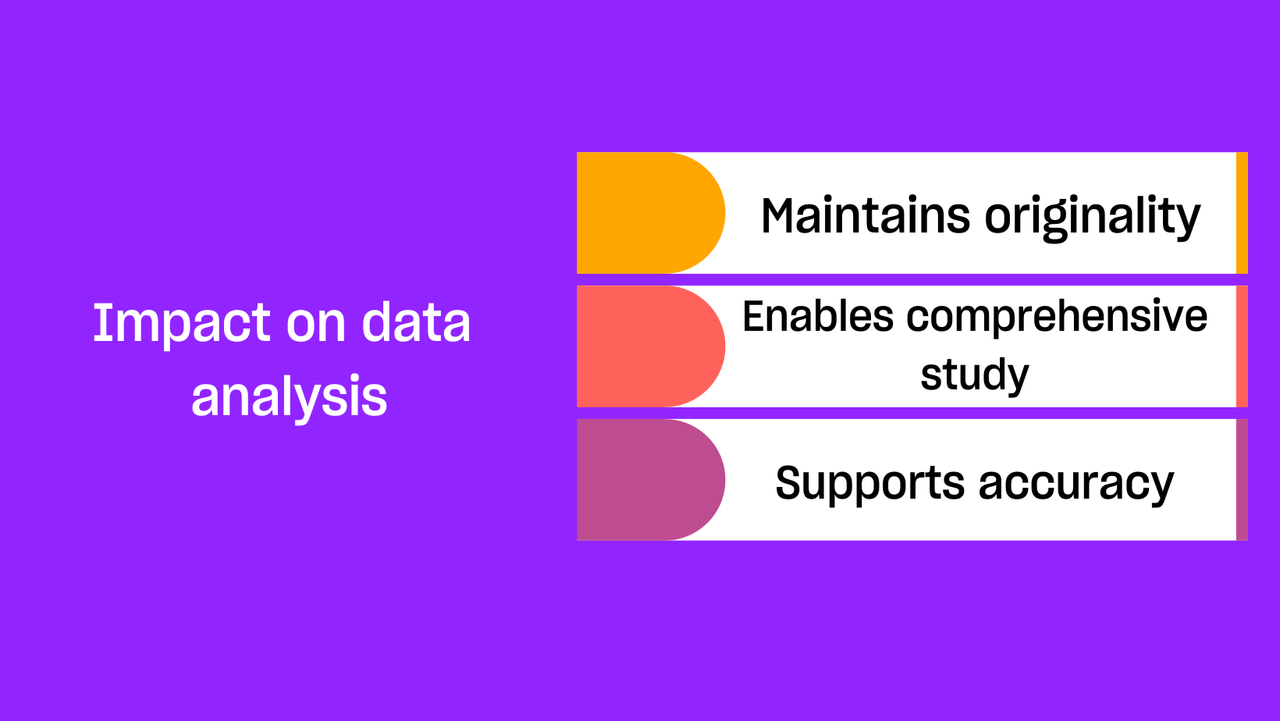
- Maintains originality: Preserving the original context of spoken words is ensured via precise transcription. For accurate interpretation of the data, this is essential.
- Enables comprehensive study: If the transcription has even minute error, it may prevent researchers from doing a thorough study of the interview data, including discourse, theme, and content analysis. Conversation analysis requires a lot of details which is possible through detailed notes of its accurate transcription.
- Supports accuracy: Data analysis in qualitative research is a very crucial step. More valid findings are produced when transcripts are accurate because they give researchers a solid foundation.
Impact on research outcomes

- Validity of findings: The reliability of the study findings is directly impacted by the quality of the transcribing. Inaccurate conclusions may result from word misinterpretation or omission.
- Reliability and reproducibility: A key component of scientific investigation is replication, which is made possible by accurate transcribing, which also increases the research's dependability.
- Reflects the voice of the participant: Accurate transcribing preserves the integrity of the participants' contributions by correctly capturing their voices.
Benefits of accurate transcription
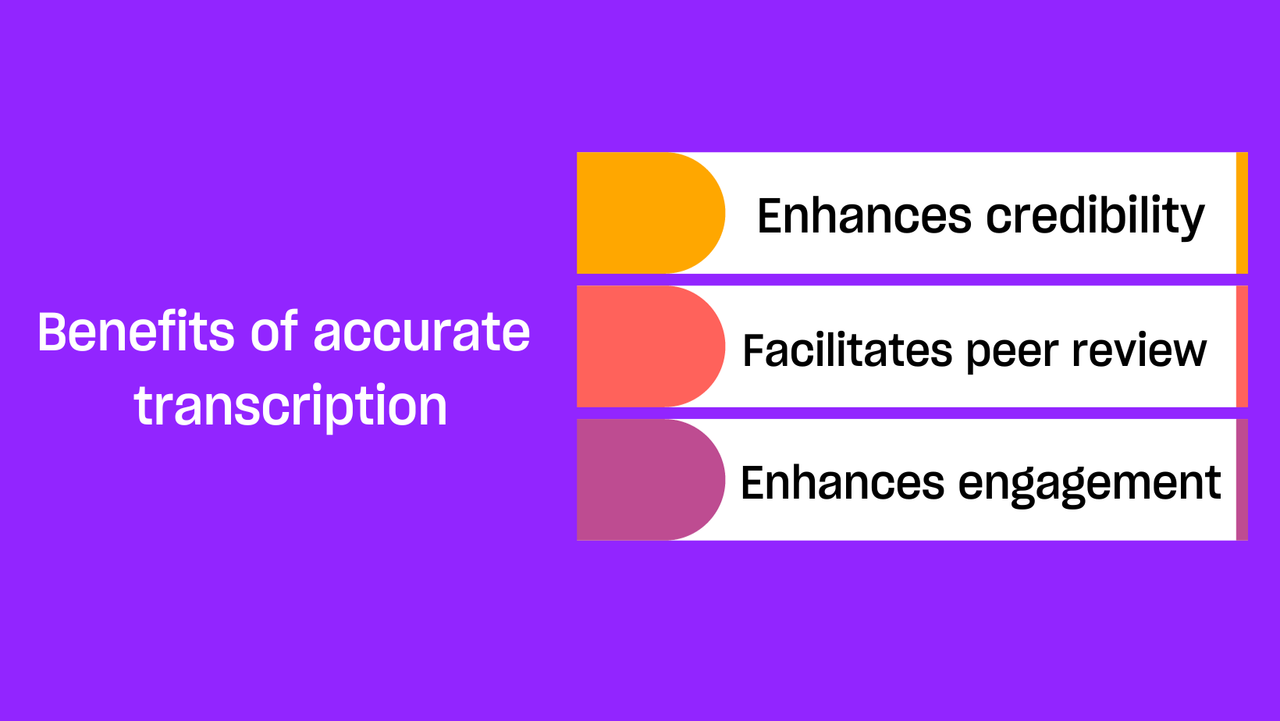
- Enhances credibility: Precisely recorded information strengthens the credibility of the study among other researchers and readers
- Facilitates peer review and cooperation: Because other researchers can comprehend and analyse the data with clarity, it makes effective peer review and cooperation possible.
- Enhances engagement with data: When data is precisely translated, researchers may interact with it at a deeper level, which results in more perceptive analysis and interpretation.
Accurate transcription plays a crucial role in maintaining the validity, reliability, and integrity of the research findings. It improves the quality and depth of data analysis, guaranteeing that the conclusions are solid, reliable, and accurate representations of the experiences and viewpoints of the participants.
Discover Good Tape’s interview transcription service
We’ve understood in depth how to transcribe interviews for qualitative research, let’s go over how you can do so accurately and quickly without having to put in much effort. Good Tape has a relatively simpler user interface which you can navigate through without any manual or instructions. Here’s what you can expect when going through the process of transcribing your audios.
- Upload your file: The first step in the process is to upload the file you need to transcribe. Make sure the file is complete and has all the information you require
- Select the language: Good Tape has a number of options when it comes to choosing the language of transcription. Select the one you want, although you can also choose the “auto-detect” option for the system to automatically identify the language in the audio.
- Transcribe the text: Once the file is uploaded and the language is chosen, proceed further by clicking the “transcribe” button. Your audio transcription process starts here.

- To wait or not to wait: If you’re a casual plan user, you will have to wait for some time for your transcription to be completed due to excessive load by the users. However, if you’re a professional or a team user, you get your results ASAP! The wait time depends on the plan you’re subscribed to .
- Get notified: You will receive a notification once your transcribed document is ready. An e-mail will be sent to your inbox containing the link to access and download the document.
Looking for a good transcribing interviews qualitative research service? Try out Good Tape’s audio-to-text transcription service today and increase your work productivity. Their AI incorporated technology makes sure that every verbal and non-verbal cue is recorded, giving your qualitative data a deeper level of understanding.
More articles


Journalistic interview: How to interview someone for an article

From text to context: A complete guide to qualitative data analysis

What is verbatim transcription?

The essential transcription services for qualitative research
We believe everyone should have access to top-quality automatic trancriptions.
That's why Good Tape is completely free to use . No credit card required.
Why is it free?
We Trust in Human Precision
20,000+ Professional Language Experts Ready to Help. Expertise in a variety of Niches.
- API Pricing
- Cost estimate
- Customer loyalty program
- Educational Discount
- Non-Profit Discount
- Green Initiative Discount1
Value-Driven Pricing
Unmatched expertise at affordable rates tailored for your needs. Our services empower you to boost your productivity.
- Special Discounts
- Enterprise transcription solutions
- Enterprise translation solutions
- Transcription/Caption API
- AI Transcription Proofreading API
Trusted by Global Leaders
GoTranscript is the chosen service for top media organizations, universities, and Fortune 50 companies.
- API solutions
GoTranscript
One of the Largest Online Transcription and Translation Agencies in the World. Founded in 2005.

Optimizing Qualitative Data Analysis: Transcription Techniques for Researchers

In the world of marketing research, qualitative data serves as a gold mine of insights, revealing the why and how behind consumer behaviors, preferences, and attitudes. However, extracting meaningful information from interviews, focus groups, and other qualitative data sources can be challenging without the right approach. This is where transcription comes into play, acting as a bridge between raw data and actionable insights. In this blog post, we delve into transcription techniques that can optimize the analysis of qualitative data, focusing on verbatim versus non-verbatim transcription and the use of timestamps.
Verbatim Transcription: Capturing Every Detail
Verbatim transcription involves transcribing exactly what is said, including all utterances, non-verbal communication cues like laughter or sighs, and false starts. This technique is invaluable when the manner in which something is said is as important as the content itself. It provides a detailed account of the conversation, offering deep insights into the participants' emotional states, hesitations, and emphasis. Researchers in fields where linguistic nuances and emotional expressions play a crucial role, such as psychology, sociology, and anthropology, often rely on verbatim transcriptions to capture the full depth of qualitative data.
Non-Verbatim Transcription: Focusing on Content
Non-verbatim transcription, also known as intelligent or clean transcription, focuses on the essence of the spoken words, omitting fillers, repetitions, and non-linguistic sounds. This approach is best suited for projects where the clarity of information is paramount, and the emotional undertones are less critical. It results in a more readable and concise document, enabling researchers to quickly identify themes and patterns without sifting through extraneous details. Non-verbatim transcription is often preferred in market research, policy analysis, and educational research, where the goal is to streamline data analysis and facilitate the identification of actionable insights.
The Role of Timestamps in Transcription
Regardless of the transcription approach chosen, timestamps are a valuable addition, providing a way to locate specific parts of the audio or video material quickly. They enable researchers to reference exact moments in the recording when analyzing or presenting findings, enhancing the accuracy and reliability of qualitative analysis. Timestamps are particularly useful in long interviews or focus groups where returning to a specific segment can be like finding a needle in a haystack without them.
Choosing the Right Transcription Technique
The choice between verbatim and non-verbatim transcription and whether to include timestamps depends on the research objectives, the nature of the qualitative data, and the intended analysis methods. Verbatim transcription is the go-to option for studies focused on communication patterns, emotional responses, and detailed content analysis. In contrast, non-verbatim transcription suits projects prioritizing efficiency and the distillation of key themes and insights.
For researchers, optimizing qualitative data analysis through transcription involves not just choosing the right technique but also ensuring accuracy and consistency across transcriptions. The use of professional transcription services can be a valuable investment, providing high-quality transcripts that faithfully represent the recorded material, thereby laying a solid foundation for insightful, data-driven conclusions.
Transcription is more than just a methodical conversion of audio into text; it's a strategic choice that can significantly impact the quality of qualitative data analysis. By carefully selecting between verbatim and non-verbatim transcription and effectively using timestamps, researchers can enhance their analysis, uncover deeper insights, and draw more nuanced conclusions from their qualitative studies. In the ever-evolving field of marketing research, adopting the right transcription techniques is crucial for turning qualitative data into meaningful strategies and decisions.
- For Customers
- For Clickworkers
- Artificial Intelligence (AI)
- clickworker News
- Crowdsourcing
- Digital Marketing
- Tips and Tricks
Table of Contents
What is Data Transcription?
What is qualitative data transcription.
- Applications
Benefits of using a Data Transcription Service
Qualitative data transcription – ultimate guide – definition, types and services.
Robert Koch
I write about AI, SEO, Tech, and Innovation. Led by curiosity, I stay ahead of AI advancements. I aim for clarity and understand the necessity of change, taking guidance from Shaw: 'Progress is impossible without change,' and living by Welch's words: 'Change before you have to'.

If you have ever conducted an interview or focus group, you know that the process of collecting data can be time-consuming. Once the data is collected, it then needs to be transcribed so that it can be analyzed. This is where qualitative data transcription comes in. It allows researchers to analyze and interpret the data more easily. There are different types of qualitative data transcription, each with its own benefits and drawbacks. This guide will help you understand the different types of transcription so that you can choose the right one for your project.
Table of Contents What is Data Transcription? What is Qualitative Data Transcription? Types Techniques Applications Challenges Benefits of using a Data Transcription Service Conclusion FAQs
Data transcription is the process of converting data from one form to another. This involves changing data from an analog source, such as paper documents or audio recordings, into a digital format, such as text or a spreadsheet. Through data transcription, data can be made more accessible, easier to use, and searchable, allowing for faster and simpler data analysis. Additionally, it can reduce the costs associated with storing physical documents or recordings.
Qualitative data transcription is converting audio and visual data into written form. It involves making interpretive decisions about what level of detail to include and how to represent the data accurately. It is the first step in qualitative data analysis and is critical for accurately capturing and interpreting the information. Qualitative data transcription can help researchers understand phenomena such as health, illness, healthcare encounters, and consumer decisions or preferences.
What is the Difference between Data Transcription and Qualitative Data Transcription?
Data transcription and qualitative data transcription are two different processes. Data transcription involves transferring recorded information from audio or video sources into written form, while qualitative data transcription involves transforming spoken language into written text. Qualitative data transcription is also more focused on capturing a person’s meaning and intent, rather than simply transferring what is said verbatim. Qualitative data transcription requires the transcriber to make subjective decisions throughout the process, such as omitting unnecessary information, correcting mistakes, and editing grammar and repetitions. This allows the transcript to more closely reflect the interviewee’s intended message. In contrast, data transcription is mainly focused on accurately transferring what is said and does not require the same level of interpretation.
What are the different Types of Data Transcription?
Transcription is the process of converting audio recordings or written documents into a written or digital format. It is an important part of many research and data collection processes. There are two main types of transcription: verbatim and edited. Verbatim transcription involves the direct transcribing of the audio, with all pauses, words, tones, and other elements of the recording included. Edited transcription will involve editing the transcript to create a more coherent and readable version.
Types of Data Transcription:
- Audio transcription: Spoken words into text format
- Video transcription: Spoken words and non-verbal cues into text format
- Handwritten transcription: Handwritten notes or documents into digital text format
- Medical transcription: Medical dictations or recordings into written reports
- Legal transcription: Legal dictations or recordings into written reports
- Business transcription: Business-related dictations or recordings into written reports
- Conference transcription: Conference or seminar recordings into text format
- Podcast transcription: Podcast episodes into text format
- Academic transcription: Lectures, interviews, or research data into text format
Types of Qualitative Data Transcription:
- Verbatim transcription: capturing every single word, sound, and utterance in the audio or video recording, including fillers, hesitations, and non-verbal cues
- Discourse transcription: focusing on the overall structure and meaning of the conversation or discussion, while omitting non-verbal cues and redundancies
- Content transcription: summarizing the key themes and points in the audio or video recording, often used in research or academic settings
- Intelligent verbatim transcription: retaining the meaningful content of the recording, while removing fillers, repetitions, and other non-essential elements
- Edited transcription: condensing and rearranging the content of the recording to create a more concise and coherent representation of the original material
Tip: Data Service for Research & Development We have a vast pool of experienced and qualified Clickworkers from all over the world who are rigorously selected and trained to ensure high-quality transcription. Our platform also utilizes advanced technology to improve accuracy and efficiency, resulting in quick turnaround times and competitive pricing. Moreover, our quality assurance process involves multiple levels of review to ensure that our clients receive the best possible service. With our commitment to excellence and client satisfaction, clickworker is the perfect partner for all your data transcription needs. Data Service
The Importance of Accuracy in Qualitative Data Transcription
Accuracy is crucial for several reasons, including:
- Data integrity: Accurate transcription ensures that the data collected from the recording is reliable and trustworthy. This is especially important in research, academic, and legal settings where the accuracy of the transcription affects the quality of the analysis or decision-making based on that data.
- Meaningful interpretation: Transcribed data is often used for analysis and interpretation. Inaccurate transcription may lead to incorrect conclusions, which can have serious implications in fields like medical research or legal proceedings.
- Credibility: The credibility of the transcription reflects the credibility of the author or the organization behind it. Inaccuracies in the transcription can lead to mistrust and discredit the entire research or project.
- Time and cost savings: Inaccurate transcription may result in additional time and costs for re-transcription or corrections. Ensuring accuracy from the start can help save time and resources in the long run.
- Ethical considerations: In some cases, inaccurate transcription can lead to misrepresentation or distortion of the recorded material, which may be considered unethical.
In summary, the importance of accuracy in qualitative data transcription cannot be overstated. It impacts the reliability, credibility, and validity of the data collected, as well as the interpretation and analysis of that data. Related: Whitepaper – Achieving AI ROI Through Data Quality and Diversity
Differences between Data Transcription and Data Entry
These two are related, but distinct processes. Here are the differences:
| Aspect | Data Entry | Data Transcription |
|---|---|---|
| Input Method | Inputting data from physical or digital sources into a database or spreadsheet | Converting audio or video recordings into text format |
| Purpose | Administrative or business purposes, such as , analysis, or reporting | Research or academic purposes, such as transcribing interviews, focus groups, or lectures |
| Skillset | Typing skills and attention to detail | Listening skills, language proficiency, and the ability to distinguish and transcribe different speakers or accents |
| Complexity | Straightforward tasks that involve inputting data into pre-defined fields or categories | More complex tasks that require the transcriber to capture not only the words spoken but also non-verbal cues and contextual information |
| Quality Control | Quality control checks to ensure accuracy and completeness of data | Higher level of quality control, including proofreading and editing, to ensure accuracy, completeness, and consistency of the transcribed text |
In summary, while data entry and data transcription share some similarities in terms of inputting data into a digital format, they are different processes with distinct purposes, skillsets, and quality control measures.
Techniques for Data Transcription
Data transcription is a process of transforming data from analog to digital form. It can be done manually by professional transcribers or automated by intelligent data processing tools. Manual data transcription is suitable for small and medium-scale businesses while automated data transcription is more suitable for large-scale businesses with huge amounts of data. Regardless of the technique used, it is important to develop a standard transcription template and provide clear instructions to ensure uniformity and accuracy.
Manual Transcription Techniques
Manual data transcription techniques refer to the process of manually transcribing data from one format to another. This can include tasks such as copying information from physical documents to digital files, transcribing audio recordings to text, or entering data from one software program into another. While there are many automated tools available for data transcription, manual transcription techniques are still necessary in many cases, particularly when dealing with handwritten documents or recordings with poor audio quality.
Here are some examples of manual data transcription techniques:
- Typing: Manually typing out information from one format into another, such as from a physical document into a digital file.
- Copying: Physically copying information from one document to another, such as copying data from a ledger book into a spreadsheet.
- Audio transcription : Listening to an audio recording and typing out what is being said in a separate document.
- Video transcription: Watching a video recording and typing out the dialogue or captions in a separate document.
- Handwritten transcription: Transcribing information from a handwritten document into a digital format, such as typing out a letter from a handwritten note.
Automated Transcription Techniques
While manual data transcription techniques are still useful in many situations, automated data transcription techniques have become increasingly popular due to their speed and efficiency. These tools can quickly transcribe large amounts of data, saving time and reducing the risk of human error. Some examples of automated data transcription techniques include:
- Optical Character Recognition (OCR): This involves using software to scan and recognize text from physical documents, such as invoices or receipts, and converting it into a digital format.
- Speech Recognition : This involves using software to transcribe spoken words into text, such as transcribing a speech or recording a conversation.
- Video Captioning: This involves using software to automatically generate captions or subtitles for video content, making it more accessible to viewers.
- Text-to-Speech : This involves using software to convert written text into spoken words, which can be useful for creating audio content or making written content more accessible.
- Data Extraction : This involves using software to automatically extract data from structured documents, such as invoices or contracts, and inputting it into a database or spreadsheet.
Applications of Data Transcription
According to a report by Grand View Research , the global transcription services market size was valued at USD 31.9 billion in 2020 and is expected to grow at a compound annual growth rate (CAGR) of 5.6% from 2021 to 2028.
Data transcription has numerous applications across various industries. Here are some examples:
- Legal: Transcription is commonly used in legal settings to transcribe depositions, court proceedings, and other legal documents.
- Healthcare : Transcription is used to convert medical records, notes, and dictations into digital formats for easy access and storage.
- Media and Entertainment: Transcription is used to generate captions and subtitles for video content, transcribe interviews and podcasts, and convert written content into audio formats.
- Education: Transcription is used to transcribe lectures, meetings, and presentations, making them more accessible to students.
- Business: Transcription is used to transcribe meetings, interviews, and phone calls for record-keeping and analysis, and to convert written content into different formats for marketing purposes.
Qualitative Data Transcription in Research
In market research, data transcription plays a crucial role in analyzing qualitative data such as focus group discussions, interviews, and surveys. Researchers can use transcription services to convert audio or video recordings into text format, allowing them to easily analyze the data and identify trends and insights.
Transcription also helps researchers to save time and reduce errors in their analysis. By transcribing data, researchers can quickly search for keywords and phrases, categorize responses, and identify patterns in the data. This can help them to draw more accurate conclusions and make better-informed decisions based on the insights gained from the data.
Furthermore, transcription can be used to transcribe customer feedback from social media or online review platforms, which can provide valuable insights into customer behavior and preferences. With the help of data transcription, market researchers can extract valuable information from a variety of sources and use it to inform business decisions, marketing strategies, and product development. Read More about the most common data analysis methods in market research .
Challenges and Solutions in Qualitative Data Transcription for Research
Data transcription is a critical part of qualitative research, as it allows researchers to accurately capture the spoken data from interviews, focus groups and other audio or video recordings. However, transcription can be an arduous and time-consuming task, as it requires careful listening, interpretation and judgements about what to include or omit. Transcribers must also be highly attentive to detail, especially when it comes to the representation of data.
In order to overcome these challenges, researchers should consider the use of transcription software, which can assist in streamlining the transcription process. Such software can help to automate certain tasks, such as transcribing pauses, detecting and correcting mistakes, and providing precise time stamps. Additionally, researchers should seek out experienced transcribers who are familiar with different levels of detail and can provide high-quality transcripts. Furthermore, researchers should have a clear idea of what level of detail is necessary and appropriate when transcribing qualitative data.
Finally, researchers should take extra precautions when dealing with data transcription in different languages. This can be done by double-checking translations and ensuring that the transcribed data accurately reflect the original audio or video recording. With the right tools and approach, researchers can successfully overcome the challenges related to transcribing qualitative data.
Outsourcing vs In-House Transcription
When it comes to transcribing qualitative research data, there is an important distinction to be made between outsourcing and in-house transcription. While the researcher who undertakes the transcription themselves can assume an inherent coherence between the research approach and approach to transcription, more care must be taken if they outsource transcription to a third-party. Outsourcing to a research assistant or commercial transcription company requires detailed and thorough instructions about the level of verbatim, style, and formatting that the researcher is expecting. It is also important to keep in mind that the transcriber should be familiar with the subject matter and have knowledge of the accents or dialects of the speakers.
In addition, there are advantages to including the transcriber in the research process. For instance, if they are present during the interview, the transcriber can gain important contextual information and observations that may not be evident in the written transcript. Finally, if the researcher chooses to outsource, it is helpful to encourage the transcriber to keep memos of their transcription process and contextual observations.
Overall, when transcribing qualitative data, researchers should consider their options for outsourcing vs in-house transcription, and create a process that meets their needs and ensures the accuracy and quality of the transcription.
Data transcription can be a time-consuming and labor-intensive task that requires a high level of accuracy. For businesses and organizations that deal with large volumes of data on a daily basis, using a data transcription service can provide numerous benefits:
- Increased accuracy: Professional transcriptionists are trained to transcribe audio and video content accurately, minimizing errors that can occur with in-house transcription.
- Time-saving: Outsourcing transcription work to a service provider frees up valuable time for businesses and organizations to focus on core activities.
- Cost-effective: By outsourcing transcription work, businesses can save on overhead costs associated with in-house transcription, such as equipment, software, and staff.
- Improved productivity: With a data transcription service, businesses can transcribe large volumes of content quickly and efficiently, improving overall productivity.
- Flexibility: Data transcription services offer a range of options, including different turnaround times, file formats, and additional services, to meet the needs of different businesses and organizations.
- Access to expertise: Professional transcriptionists have experience transcribing a wide range of content types, including legal, medical, and technical content, ensuring high-quality results for businesses and organizations in different industries.
How to choose the right Data Transcription Service for your needs
Choosing the right data transcription service can be daunting, especially with so many available options. When selecting a transcription service, it’s important to consider factors such as turnaround time, accuracy, cost, and security.
Firstly, consider the turnaround time offered by the service provider. If you have a tight deadline, you may need a service that can provide a faster turnaround time.
Secondly, accuracy is essential when it comes to transcription. You want to choose a service that provides accurate transcriptions, especially if the data is being used for research or legal purposes.
Thirdly, the cost is an important consideration. Different transcription services have different pricing models, so it’s important to choose one that fits your budget.
Lastly, consider the security measures provided by the transcription service. The service should have secure data transfer protocols, data encryption, and confidentiality agreements in place to protect your data.
By considering these factors, you can choose the right transcription service that meets your needs and provides accurate, timely, and secure transcription services.
In conclusion, data transcription is an important step in the research project process. It requires the right equipment, attention to detail, and an understanding of the context of the data. There are a variety of ways this can be accomplished, from manually transcribing onto a word-processing document to utilizing specific tools like foot pedals and browser add-ons. Re-reading the transcripts and conducting member checking can help ensure the accuracy of the transcripts. After transcribing is complete, researchers are able to move on to data analysis, which will help provide insights into their research project. Ultimately, the aim of data transcription is to accurately capture the data in order to reach meaningful conclusions about the project.
FAQs on Data Transcription
What is transcribed data.
Transcribed data is the text that results from having audio or video recordings converted into a written format. This can be done by either typing out the recording verbatim, or by taking notes while listening to or watching the recording. The written transcript can then be used for a variety of purposes, such as creating subtitles or providing searchable text.
What is a data transcription analysis?
A data transcription analysis is the process of analyzing a set of data in order to extract useful information. This can be done manually or through automated means. Data transcription analysis is often used in the fields of research, data mining, and business intelligence.
What is qualitative interview transcription?
Qualitative interview transcription is the process of converting an audio recording of an interview into text. This can be useful for analyzing interviews for research purposes, or for creating written records that can be shared with others.
Related Posts
- How to Transcribe Audio to Text: A Guide to Audio Transcription and Speech Recognition
- What is Primary Data Collection? Types, Advantages, and Disadvantages
- Solutions Overview
- Audio Datasets & Voice Datasets
- Image Datasets & Photo Datasets
- Video Datasets
- Image Annotation
- Product Categorization & Tagging
- Image & Video Tagging
- Sentiment Analysis
- Video Analysis
- Search Relevance
- Customer FAQ
- About Crowdsourcing
- Crowdsourcing Glossary
- Content Marketing Glossary
- AI Glossary
- Clickworker Job
- Clickworker FAQ
- Clickworker Registration
- Clickworker App
- Clickworker Data Security
clickworker USA
2 Park Avenue, 20th Floor New York, NY 10016 USA
clickworker Europe
Büropark Bredeney Theodor-Althoff-Str. 41 45133 Essen, Germany

- Cookie Declaration
- Strictly Necessary Cookies
- Additional Cookies
This website uses cookies to provide you with the best user experience possible. Cookies are small text files that are cached when you visit a website to make the user experience more efficient. We are allowed to store cookies on your device if they are absolutely necessary for the operation of the site. For all other cookies we need your consent.
You can at any time change or withdraw your consent from the Cookie Declaration on our website. Find the link to your settings in our footer.
Find out more in our privacy policy about our use of cookies and how we process personal data.
Necessary cookies help make a website usable by enabling basic functions like page navigation and access to secure areas of the website. The website cannot properly operate without these cookies.
If you disable this cookie, we will not be able to save your preferences. This means that every time you visit this website you will need to enable or disable cookies again.
Any cookies that may not be particularly necessary for the website to function and is used specifically to collect user personal data via analytics, ads, other embedded contents are termed as additional cookies.
Please enable Strictly Necessary Cookies first so that we can save your preferences!

- Onsite training
3,000,000+ delegates
15,000+ clients
1,000+ locations
- KnowledgePass
- Log a ticket
01344203999 Available 24/7
Transcription vs. Translation: Discover the Differences
Transcription and Translation services involve language conversion in the world of linguistics and communication. However, they serve different purposes and cater to different needs. Read this blog to learn everything about Transcription vs Translation and how Translation converts text between languages and Transcription.

Exclusive 40% OFF
Training Outcomes Within Your Budget!
We ensure quality, budget-alignment, and timely delivery by our expert instructors.
Share this Resource
- Active Listening Training
- Voice Over Artist Course
- Audio Transcription Course
- Speed Writing Course
- Introduction to Podcasting

Imagine trying to understand a foreign language film without subtitles or attempting to capture every word of an important speech without a written record. This is where Transcription and Translation come into play. These processes are pivotal in converting spoken words into written text and translating text between languages, respectively. However, what are the differences between Transcription vs Translation?
Understanding the differences between Transcription vs Translation is essential for anyone looking to improve communication and accessibility. This blog will guide you through their unique roles, methods, and how they contribute to effective communication across different mediums and languages.
Table of Contents
1) What is Transcription?
2) What is Translation?
3) Difference Between Transcription and Translation
4) How do Transcription and Translation Work?
5) Conclusion
What is Transcription?
Transcription is the process of converting spoken language into written text. It is applicable to various mediums such as audio and video recordings, even live interactions. There are two main types: Verbatim and non-verbatim.
Verbatim transcription captures every word exactly as spoken. This comprehensive approach includes all sounds and is essential for full accessibility, particularly for individuals who are Deaf or have hearing difficulties, as it allows complete participation in video content.
Non-verbatim transcription involves the content to highlight the main points. This can be practical for certain situations. However, it may not provide the same level of accessibility as verbatim transcriptions.

Types of Transcription
There are basically four types of transcription: edited, verbatim, intelligent, and phonetic. Each has its own positives and negatives. Let’s learn about it in detail below.

1) Edited Transcription
This transcription style is also known as clean verbatim, which refines the source material for enhanced readability and formality. It involves meticulous editing to correct grammatical errors and remove superfluous elements like slang or incomplete sentences. The result is a polished document that retains the essence of original message but may adopt a more formal tone.
Edited transcription is particularly beneficial in professional environments like business. There, it is commonly used for documenting conferences, seminars, and speeches. Its clarity and formal tone make it suitable for academic and medical records, which often require precision and readability. Additionally, this transcription type is advantageous in publishing and translation, where the polished documents are intended for a wider audience.
2) Intelligent Transcription
Serving as a bridge between edited and verbatim, this style focuses on natural readability. It lightly edits for grammar, excludes redundant phrases and fillers, and streamlines speech without altering the message’s essence. The key lies in capturing the core meaning while keeping the original tone largely intact. Suitable for business and medical contexts where full formality isn’t required, it ensures the main ideas are clearly communicated.
3) Verbatim Transcription
This comprehensive transcription method captures every aspect of the audio or video source, including verbal nuances and ambient sounds. It’s a meticulous process that documents all details, such as speech patterns, emotional cues, and environmental noises. Verbatim transcription is crucial in legal contexts where the authenticity of dialogue and behaviour is paramount, as well as in scientific and marketing research that demands precision and accounts for every slight variation.
4) Phonetic Transcription
This transcription style is distinct because it focuses on the pronunciation of words using the International Phonetic Alphabet (IPA). It’s not about the content’s meaning but its sound. Each sound, or phoneme, is denoted by a unique symbol, capturing the nuances of speech.
Its specialised nature makes it invaluable in linguistics, language education, and speech therapy. It’s also used in the entertainment industry for accurate representation of accents and dialects. Phonetic transcription is essential for anyone studying or analysing the spoken word at a granular level.

What is Translation?
Translation is the process of converting spoken text from one language to another. It expands the video's appeal to wider audiences. There are several platforms that offer various methods for sharing Translations. This includes on-screen captions or subtitles for real-time viewing, or over-dubbing recordings to allow viewers to hear audio in their native language. Real-time Translation options extend to live communications like conference calls and seminars.
Multilingual Translations can surely enhance the reach of your video. This enables brands to connect with diverse global audiences. Investing in Translations not only boosts visibility but also supports international customers and employees. As remote work becomes more prevalent, translated communications facilitate effective and equitable interactions across regions and departments.
Types of Translation
Translation serves the essential role of bridging language barriers. There are various translation types exist, each tailored to specific needs. However, they all share the common goal of enabling cross-lingual communication. The five common translation types are:

This is the art of translating literature like plays, novels, and poems. Here, the translator's task is to convey the text's meaning and the author's stylistic flair. Literary works are rich with stylistic elements, including rhythm, meter, wordplay, and imagery that often lack direct equivalents in other languages.
Translators must be inventive to evoke the impact of the original work. They also need to navigate the cultural nuances and contextual subtleties that may seem untranslatable. A translator's skill lies in capturing the author's unique voice. They must ensure the translation reflects the original style and essence. Therefore, it requires a deep understanding of literary devices in both the source and target languages.
2) Technical Translation
This translation type is essential for global businesses to meet diverse standards. It includes a vast array of documents, from patents to MSDS, across sectors like engineering and pharmaceuticals. The challenge lies in the specialised terminology unique to each field.
Effective technical translation demands not just linguistic skills but also a deep understanding of the subject matter, industry-specific jargon, and legal stipulations. A proficient technical translator ensures accurate communication by adeptly navigating these complexities.
3) Administrative Translation
This translation category is integral to business management. It helps translate documents like contracts, newsletters, and invoices that facilitate daily operations. This is important for companies to expand or sustain their international reach as it helps to dismantle language barriers and foster a diverse workforce. Such translations are key to aligning business partners, investors, and employees. It ensures unified communication for a more efficient and collaborative corporate environment.
4) Financial Translation
Essential for the global financial sector, this translation type supports banks and financial institutions in broadening service access. It helps maintain customer trust while adhering to international standards. As these entities venture into emerging markets, the need for precise financial translation surges. It encompasses a spectrum of documents, all demanding translators with specialised knowledge and an understanding of stringent regulations.
Financial Translators must grasp the intricacies of local and international laws to ensure compliance. With the financial world growing rapidly, translators must continuously update their vocabulary to keep pace with the latest industry terms and innovations.
5) Legal Translation
This intricate translation field involves converting legal documents like contracts, proposals, and court transcripts into another language. It’s important to ensure that these documents are comprehensible and enforceable across different cultures and legal systems. Legal Translators must really understand the laws and the way of life related to both languages they translate between.
This deep knowledge is important to make sure the translation is correct and keeps its legal meaning. Accuracy is critical in legal translation, as even small mistakes can have substantial legal consequences. Therefore, legal translation often requires collaboration with legal experts. This guarantees that the translated text upholds the same legal weight as the original.
Master effective communication with our Active Listening Training – Sign up today!
Difference Between Transcription and Translation
Translation and transcription share the goal of accurate content representation, but they diverge significantly in their processes. Here is a table that represents the key differences between transcription vs translation.
|
|
|
|
|
| Converts material into another language. | Converts spoken words into written form in the same language. |
|
| Often more complex due to cultural and contextual nuances. | Generally, it is less complex and involves recording the content as is. |
|
| Requires critical thinking to express sentiments accurately in a different language. | Involves typing out words exactly as they are spoken. |
|
| Must account for non-literal expressions and contextually accurate meanings. | Straightforward conversion but may include editing for clarity. |
|
| Demands a deep understanding of both the source and target languages. | Requires good listening skills and fast, accurate typing. |
How do Transcription and Translation Work?
Now that we have learned the functions of Transcription and Translation, let's take a look at how these processes actually operate. There are various approaches available when it comes to transcribing or translating video content.
Some companies opt to assign in-house staff to handle Transcription or Translation tasks. This may seem cost-effective initially. However, untrained individuals often produce inaccuracies that can compromise accessibility and the viewing experience.
Alternatively, certain media platforms provide automatic Transcription and Translation features for video content. Although convenient for brands producing large volumes of content, these AI-driven tools often fall short in accuracy, failing to meet accessibility standards.
Conclusion
Translation and Transcription both aim to transform information. However, they operate in distinct realms with specific methods, goals, and hurdles. Translation facilitates cross-cultural communication and knowledge transfer by bridging linguistic gaps. Meanwhile, Transcription serves practical functions like documentation, research, and accessibility. Understanding the disparities between "Transcription vs Translation" is vital for appreciating their unique methodologies and roles in preserving human understanding.
Do you want to understand the five vocal elements for voice training and different voice accents? Sign up for our Voice Over Artist Course today!
Frequently Asked Questions
Translation and Transcription services bridge linguistic gaps. They contribute to cross-cultural communication and understanding by making content accessible to diverse audiences.
Commonly translated or transcribed languages include English, Spanish, Mandarin, and Arabic. The approach may differ based on language complexity, script, and cultural context.
The Knowledge Academy takes global learning to new heights, offering over 30,000 online courses across 490+ locations in 220 countries. This expansive reach ensures accessibility and convenience for learners worldwide.
Alongside our diverse Online Course Catalogue, encompassing 17 major categories, we go the extra mile by providing a plethora of free educational Online Resources like News updates, Blogs , videos, webinars, and interview questions. Tailoring learning experiences further, professionals can maximise value with customisable Course Bundles of TKA .
The Knowledge Academy’s Knowledge Pass , a prepaid voucher, adds another layer of flexibility, allowing course bookings over a 12-month period. Join us on a journey where education knows no bounds.
The Knowledge Academy offers various Personal Development Courses , including Translation Course Stress Management and Managing Difficult Conversation Training. These courses cater to different skill levels, providing comprehensive insights into Effective Communication .
Our Business Skills Blogs cover a range of topics related to Personal Development, offering valuable resources, best practices, and industry insights. Whether you are a beginner or looking to advance your Translation skills, The Knowledge Academy's diverse courses and informative blogs have you covered.
Upcoming Business Skills Resources Batches & Dates
Fri 21st Jun 2024
Fri 2nd Aug 2024
Fri 18th Oct 2024
Fri 20th Dec 2024
Get A Quote
WHO WILL BE FUNDING THE COURSE?
My employer
By submitting your details you agree to be contacted in order to respond to your enquiry
- Business Analysis
- Lean Six Sigma Certification
Share this course
Our biggest spring sale.

We cannot process your enquiry without contacting you, please tick to confirm your consent to us for contacting you about your enquiry.
By submitting your details you agree to be contacted in order to respond to your enquiry.
We may not have the course you’re looking for. If you enquire or give us a call on 01344203999 and speak to our training experts, we may still be able to help with your training requirements.
Or select from our popular topics
- ITIL® Certification
- Scrum Certification
- Change Management Certification
- Business Analysis Courses
- Microsoft Azure Certification
- Microsoft Excel Courses
- Microsoft Project
- Explore more courses
Press esc to close
Fill out your contact details below and our training experts will be in touch.
Fill out your contact details below
Thank you for your enquiry!
One of our training experts will be in touch shortly to go over your training requirements.
Back to Course Information
Fill out your contact details below so we can get in touch with you regarding your training requirements.
* WHO WILL BE FUNDING THE COURSE?
Preferred Contact Method
No preference
Back to course information
Fill out your training details below
Fill out your training details below so we have a better idea of what your training requirements are.
HOW MANY DELEGATES NEED TRAINING?
HOW DO YOU WANT THE COURSE DELIVERED?
Online Instructor-led
Online Self-paced
WHEN WOULD YOU LIKE TO TAKE THIS COURSE?
Next 2 - 4 months
WHAT IS YOUR REASON FOR ENQUIRING?
Looking for some information
Looking for a discount
I want to book but have questions
One of our training experts will be in touch shortly to go overy your training requirements.
Your privacy & cookies!
Like many websites we use cookies. We care about your data and experience, so to give you the best possible experience using our site, we store a very limited amount of your data. Continuing to use this site or clicking “Accept & close” means that you agree to our use of cookies. Learn more about our privacy policy and cookie policy cookie policy .
We use cookies that are essential for our site to work. Please visit our cookie policy for more information. To accept all cookies click 'Accept & close'.
- Search Menu
- Sign in through your institution
- Advance Articles
- Editor's Choice
- Supplements
- Spotlight Issues
- Image Gallery
- ESC Journals App
- ESC Content Collections
- Author Guidelines
- Submission Site
- Why publish with CVR?
- Open Access Options
- Read & Publish
- Author Resources
- Self-Archiving Policy
- About Cardiovascular Research
- About the European Society of Cardiology
- ESC Publications
- Journal Career Network
- Editorial Board
- ESC membership
- Advertising and Corporate Services
- Developing Countries Initiative
- Dispatch Dates
- Terms and Conditions
- Journals on Oxford Academic
- Books on Oxford Academic
Article Contents
Data availability, laminin: guardian against dna damage by transcription stress.
The opinions expressed in this article are not necessarily those of the Editors of Cardiovascular Research or of the European Society of Cardiology.
Conflict of interest: none declared.
- Article contents
- Figures & tables
- Supplementary Data
James W S Jahng, Joseph C Wu, Laminin: guardian against DNA damage by transcription stress, Cardiovascular Research , 2024;, cvae122, https://doi.org/10.1093/cvr/cvae122
- Permissions Icon Permissions
This editorial refers to ‘DNA double-stranded breaks, a hallmark of aging, defined at the nucleotide resolution, are increased and associated with transcription in the cardiac myocytes in LMNA-cardiomyopathy’, by B. Cathcart et al ., https://doi.org/10.1093/cvr/cvae063 .
The human genome spans 3.2 billion base pairs, and more than 20 000 genes govern various cellular processes and functions. A linear DNA is wrapped around histones to form nucleosomes that are further compacted into chromatin fibres. Chromatin fibres undergo a series of additional compression and condensation to form various three-dimensional structural units such as chromatin loops, topologically associated domains, and chromosome territory. Based on the gene activity, chromatin fibres can be grouped into two compartments: A and B compartments. A compartments generally contain transcriptionally active euchromatin and are found inside the nucleus. On the other hand, B compartments contain inactive heterochromatin and are located at the nuclear periphery. 1 At the nuclear periphery, the inner side of nuclear envelop is coated by nuclear lamina that contains a complex of protein and nuclear lamins, members of type V intermediate filament, which play important roles in nuclear processes and mechanical properties of nuclei.
There are two types of lamins, A type and B type. A-type lamins include lamins A/C and are transcribed from a single gene called LMNA, whereas B-type lamins include lamins B1 and B2 that are transcribed from two separate genes, LMNB1 and LMNB2. Cells lacking lamins or expressing mutant forms of lamins fail to establish a proper nuclear lamina, resulting in laminopathies that is characterized by nuclear distortion, aberrant gene expression, and impaired mechanotransduction. Laminopathies are associated with various diseases such as the premature aging syndrome, including Hutchinson–Gilford progeria syndrome, restrictive dermopathy, lipodystrophy, and muscular dystrophy. Notably, mutations in LMNA lead to arrhythmogenic and/or dilated cardiomyopathy (DCM) in patients or animal models of LMNA cardiomyopathy. 2
In this issue of Cardiovascular Research , Cathcart et al . 3 revealed the genome-wide distribution of double stranded breaks (DSBs) in cardiomyocytes for the first time with END-sequencing (END-seq) and provided novel insights into the role of nuclear lamina in regulating baseline DSBs by transcription. END-seq is a sensitive method for detecting DSBs at nucleotide resolution by ligating a sequencing adaptor to the ends of DSBs enabling the subsequent next generation sequencing to map DSB distribution at the genome-wide level. 4 Cathcart et al . 3 performed END-seq on 1 million cardiomyocytes from a well-established LMNA-DCM model (Myh6-Cre:LmnaF/F) 5 , 6 and compared them with wild-type (WT) mice. END-seq was conducted on 3-week-old mice, close to the end of their 4-week lifespan. Here, END-seq revealed a relatively modest increase in total DSBs, approximately three times higher compared to WT (2.2% vs. 0.8%). Given severe cardiac phenotypes typically observed at 3-week-old animals in earlier studies, 5 , 6 this change may seem mild yet the spatial distribution of DSBs remains intriguing ( Figure 1 ).

Excessive DSB distribution in the genome due to transcription stress. Chromatin fibres are organized into distinct territories that can be largely classified into A and B compartments. A compartments typically reside in the nuclear interior and are composed of euchromatin with high level of gene expression. B compartments typically reside in the nuclear periphery and are composed of heterochromatin with low level of gene expression. Transcription has been known to cause genomic instability due to torsional stress by unwinding double stranded DNA, inducing transient DNA damage by TOP2. In addition, transcription results in formation of non-B-DNA structures such as G-quadruplex and Z-DNA in non-transcribed DNA strand, and excessive presence of non-B-DNA during transcription has been reported to be associated with genomic instability. The study by Cathcart et al . 3 demonstrated that cardiomyocytes from LMNA-DCM mice suffered from excessive DSBs at baseline due to transcriptional stress. Combining END-seq and CUT&RUN assay, they found that juxta-positioned LADs in LMNA-DCM mice samples had high levels of DSBs, suggesting a significant role for nuclear laminins in regulating transcriptional stress. DSBs, double stranded breaks; LADs, laminin associated domains; LMNA, laminin A; RNA polymerase II, Pol II; TOP2, topoisomerase 2; TSS, transcription start site.
Cathcart et al . 3 also performed in-depth profiling of DSB distribution by classifying the gene and the intergenic regions. By comparing RNA sequencing (RNA-seq) data from the earlier study, 5 DSBs were preferentially localized at the highly expressed gene regions sites, especially at the transcription start site. A motif analysis of the DSB sites identified 26 transcription factors, including ETS, ETV4, GATA4, and NFATC3, which are related to cardiac development and cardiac function. Additionally, the authors classified DSB sites by distinguishing canonical double helical B-DNA and non-B-DNA structures. Intriguingly, DSB sites are preferentially located near the non-B-DNA structures like Z-DNA. Given that non-B-DNA structures are known to be associated with transcriptional stress, 7 Cathcart’s team further investigated the relationship between the nuclear lamina and transcription stress. Transcription stress refers to the physical torsional stress imposed by the unwinding of double stranded DNA, transient exposure of non-transcribed strand to the extrinsic DNA damages, and secondary non-B-DNA structures, which collectively accumulate DSBs at R-loops. Cathcart et al . 3 conducted a CUT&RUN assay, a chip-seq equivalent technique with lower background, against LMNA antibody to identify ‘lamina-associated domains’ (LADs) which accounted for 30–40% of the genome. 1 The authors delineated constitutive LAD (cLAD) regions to reveal a significant reduction in DSB distribution compared to non-LAD regions. Conversely, in cardiomyocytes from LMNA-DCM mice, juxtaposed LAD regions or originally cLAD regions in WT samples exhibited excessive DSBs, suggesting that nuclear lamina can protect LADs from DSBs induced by transcriptional stress.
Taken together, the current study 3 not only confirms the conventional understanding that genes in LADs are repressed in in vivo cardiomyocytes but it also provides new insights into how aberrant transcription activations increases the burden of DSBs, which are the underlying cause of heart failure due to laminopathies. This is consistent with previous results showing that targeting subsequent pathway (cGAS-STING, TP53) or epigenetic regulator of DSBs can successfully reverse the progression of heart failure by LMNA deficiency. 5 , 6 Particularly, BRD4 inhibition to target transcription stress may be further investigated as a novel approach as it has been demonstrated effective against non-LMNA heart failure. 8
One important question raised by this study is the source of excessive DSBs. Is it due to accumulation of R-loops? Or lack of topoisomerase activity? The current study is limited in that it provides only observational evidence of DSBs accumulation at the active transcription site. Future follow-up investigations should focus on identifying the causality of DSBs accumulation through mechanistic studies. In addition, this study was performed in LMNA depletion models. To expedite the significance of LADs and the burden of DSBs by transcription stress in LMNA-DCM, future efforts could be performed in human models with clinically relevant mutations such as induced pluripotent stem cell derived cardiomyocytes. 9 , 10 Overall, this study represents the first exploration of DSB sites in in vivo heart samples that found a site-specific localization of DSBs in actively transcribed regions and LADs.
This publication was supported by grants from the National Institutes of Health R01 HL113006, R01 HL130020, and R01 HL141371 (J.C.W.). All other authors report no relationships relevant to the contents of this paper to disclose.
No new data were generated or analysed in support of this research.
Pombo A , Dillon N . Three-dimensional genome architecture: players and mechanisms . Nat Rev Mol Cell Biol 2015 ; 16 : 245 – 257 .
Google Scholar
Chen SN , Sbaizero O , Taylor MRG , Mestroni L . Lamin A/C cardiomyopathy: implications for treatment . Curr Cardiol Rep 2019 ; 21 : 160 .
Cathcart B , Cheedipudi SM , Rouhi L , Zhao Z , Gurha P , Marian AJ . DNA double-stranded breaks, a hallmark of aging, defined at the nucleotide resolution, are increased and associated with transcription in the cardiac myocytes in LMNA-cardiomyopathy . Cardiovasc Res 2024 : cvae063 . doi: 10.1093/cvr/cvae063 . Published online ahead of print 5 April 2024.
Canela A , Sridharan S , Sciascia N , Tubbs A , Meltzer P , Sleckman BP , Nussenzweig A . DNA breaks and end resection measured genome-wide by end sequencing . Mol Cell 2016 ; 63 : 898 – 911 .
Auguste G , Rouhi L , Matkovich SJ , Coarfa C , Robertson MJ , Czernuszewicz G , Gurha P , Marian AJ . BET bromodomain inhibition attenuates cardiac phenotype in myocyte-specific lamin A/C-deficient mice . J Clin Invest 2020 ; 130 : 4740 – 4758 .
Cheedipudi SM , Asghar S , Marian AJ . Genetic ablation of the DNA damage response pathway attenuates lamin-associated dilated cardiomyopathy in mice . JACC Basic Transl Sci 2022 ; 7 : 1232 – 1245 .
Kim N , Jinks-Robertson S . Transcription as a source of genome instability . Nat Rev Genet 2012 ; 13 : 204 – 214 .
Edwards DS , Maganti R , Tanksley JP , Luo J , Park JJH , Balkanska-Sinclair E , Ling J , Floyd SR . BRD4 prevents R-loop formation and transcription–replication conflicts by ensuring efficient transcription elongation . Cell Rep 2020 ; 32 : 108166 .
Lee J , Termglinchan V , Diecke S , Itzhaki I , Lam CK , Garg P , Lau E , Greenhaw M , Seeger T , Wu H , Zhang JZ , Chen X , Gil IP , Ameen M , Sallam K , Rhee J-W , Churko JM , Chaudhary R , Chour T , Wang PJ , Snyder MP , Chang HY , Karakikes I , Wu JC . Activation of PDGF pathway links LMNA mutation to dilated cardiomyopathy . Nature 2019 ; 572 : 335 – 340 .
Sayed N , Liu C , Ameen M , Himmati F , Zhang JZ , Khanamiri S , Moonen JR , Wnorowski A , Cheng L , Rhee JW . Clinical trial in a dish using iPSCs shows lovastatin improves endothelial dysfunction and cellular cross-talk in LMNA cardiomyopathy . Sci Transl Med 2020 ; 12 : eaax9276 . doi: 10.1126/scitranslmed.aax9276 .
Author notes
| Month: | Total Views: |
|---|---|
| June 2024 | 88 |
Email alerts
- DNA double-stranded breaks, a hallmark of aging, defined at the nucleotide resolution, are increased and associated with transcription in the cardiac myocytes in LMNA-cardiomyopathy
Citing articles via
- Recommend to Your Librarian
- Journals Career Network
Affiliations
- Online ISSN 1755-3245
- Print ISSN 0008-6363
- Copyright © 2024 European Society of Cardiology
- About Oxford Academic
- Publish journals with us
- University press partners
- What we publish
- New features
- Open access
- Institutional account management
- Rights and permissions
- Get help with access
- Accessibility
- Advertising
- Media enquiries
- Oxford University Press
- Oxford Languages
- University of Oxford
Oxford University Press is a department of the University of Oxford. It furthers the University's objective of excellence in research, scholarship, and education by publishing worldwide
- Copyright © 2024 Oxford University Press
- Cookie settings
- Cookie policy
- Privacy policy
- Legal notice
This Feature Is Available To Subscribers Only
Sign In or Create an Account
This PDF is available to Subscribers Only
For full access to this pdf, sign in to an existing account, or purchase an annual subscription.
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 05 June 2024
A disease-associated gene desert directs macrophage inflammation through ETS2
- C. T. Stankey ORCID: orcid.org/0000-0001-5710-1716 1 , 2 , 3 na1 ,
- C. Bourges ORCID: orcid.org/0000-0001-8122-0475 1 na1 ,
- L. M. Haag ORCID: orcid.org/0000-0002-3754-5317 4 na1 ,
- T. Turner-Stokes 1 , 2 ,
- A. P. Piedade 1 ,
- C. Palmer-Jones 5 , 6 ,
- I. Papa ORCID: orcid.org/0000-0003-3167-7623 1 ,
- M. Silva dos Santos ORCID: orcid.org/0000-0003-2404-8490 7 ,
- Q. Zhang 8 ,
- A. J. Cameron ORCID: orcid.org/0000-0002-7065-9033 9 ,
- A. Legrini 9 ,
- T. Zhang 9 ,
- C. S. Wood 9 ,
- F. N. New ORCID: orcid.org/0000-0001-6213-4731 10 ,
- L. O. Randzavola 2 ,
- L. Speidel 11 , 12 ,
- A. C. Brown 13 ,
- A. Hall 14 , 15 ,
- F. Saffioti ORCID: orcid.org/0000-0001-7635-9931 6 , 14 ,
- E. C. Parkes 1 ,
- W. Edwards 16 ,
- H. Direskeneli 17 ,
- P. C. Grayson 18 ,
- L. Jiang 19 ,
- P. A. Merkel 20 , 21 ,
- G. Saruhan-Direskeneli ORCID: orcid.org/0000-0002-6903-7173 22 ,
- A. H. Sawalha ORCID: orcid.org/0000-0002-3884-962X 23 , 24 , 25 , 26 ,
- E. Tombetti 27 , 28 ,
- A. Quaglia 15 , 29 ,
- D. Thorburn 6 , 14 ,
- J. C. Knight ORCID: orcid.org/0000-0002-0377-5536 13 , 30 , 31 ,
- A. P. Rochford 5 , 6 ,
- C. D. Murray 5 , 6 ,
- P. Divakar 10 ,
- M. Green 32 ,
- E. Nye 32 ,
- J. I. MacRae ORCID: orcid.org/0000-0002-1464-8583 7 ,
- N. B. Jamieson ORCID: orcid.org/0000-0002-9552-4725 9 ,
- P. Skoglund 11 ,
- M. Z. Cader 16 , 33 ,
- C. Wallace ORCID: orcid.org/0000-0001-9755-1703 16 , 34 ,
- D. C. Thomas ORCID: orcid.org/0000-0002-9738-2329 16 , 33 &
- J. C. Lee ORCID: orcid.org/0000-0001-5711-9385 1 , 5 , 6
Nature volume 630 , pages 447–456 ( 2024 ) Cite this article
146k Accesses
1039 Altmetric
Metrics details
- Autoimmunity
- Functional genomics
- Immunogenetics
Increasing rates of autoimmune and inflammatory disease present a burgeoning threat to human health 1 . This is compounded by the limited efficacy of available treatments 1 and high failure rates during drug development 2 , highlighting an urgent need to better understand disease mechanisms. Here we show how functional genomics could address this challenge. By investigating an intergenic haplotype on chr21q22—which has been independently linked to inflammatory bowel disease, ankylosing spondylitis, primary sclerosing cholangitis and Takayasu’s arteritis 3 , 4 , 5 , 6 —we identify that the causal gene, ETS2 , is a central regulator of human inflammatory macrophages and delineate the shared disease mechanism that amplifies ETS2 expression. Genes regulated by ETS2 were prominently expressed in diseased tissues and more enriched for inflammatory bowel disease GWAS hits than most previously described pathways. Overexpressing ETS2 in resting macrophages reproduced the inflammatory state observed in chr21q22-associated diseases, with upregulation of multiple drug targets, including TNF and IL-23. Using a database of cellular signatures 7 , we identified drugs that might modulate this pathway and validated the potent anti-inflammatory activity of one class of small molecules in vitro and ex vivo. Together, this illustrates the power of functional genomics, applied directly in primary human cells, to identify immune-mediated disease mechanisms and potential therapeutic opportunities.
Similar content being viewed by others
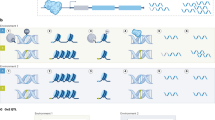
Genotype × environment interactions in gene regulation and complex traits

Genome-wide meta-analyses of restless legs syndrome yield insights into genetic architecture, disease biology and risk prediction

Plasma proteomic associations with genetics and health in the UK Biobank
Nearly 5% of humans live with an autoimmune or inflammatory disease. These heterogeneous conditions, ranging from Crohn’s disease and ulcerative colitis (collectively inflammatory bowel disease (IBD)) to psoriasis and lupus, all require better therapies, but only 10% of drugs entering clinical development ever become approved treatments 2 . This high failure rate is mainly due to a lack of efficacy 8 and reflects our poor understanding of disease mechanisms. Genetics provides a unique opportunity to address this, with hundreds of loci now directly linked to the pathogenesis of immune-mediated diseases 9 . Indeed, drugs that target pathways implicated by genetics have a far higher chance of being effective 10 .
However, to fully realize the potential of genetics, knowledge of where risk variants lie must be translated into an understanding of how they drive disease 9 . Animal models can help with this, especially for coding variants in conserved genes 11 , 12 . Unfortunately, most risk variants do not lie in coding DNA, but in less-well-conserved, non-coding genomic regions. Resolving the biology at these loci is a formidable task, as the same DNA sequence can function differently depending on the cell type and/or external stimuli 9 . Most non-coding variants are thought to affect gene regulation 13 , but difficulties identifying causal genes, which may lie millions of bases away, and causal cell types, which may only express implicated genes under certain conditions, have hindered efforts to identify disease mechanisms. For example, although genome-wide association studies (GWASs) have identified over 240 IBD risk loci 3 , including several possible drug targets, fewer than 10 have been mechanistically resolved.
Molecular mechanisms at chr21q22
Some genetic variants predispose to multiple diseases, highlighting both their biological importance and an opportunity to study shared disease mechanisms. One notable example is an intergenic region on chromosome 21q22 (chr21q22), where the major allele haplotype predisposes to five inflammatory diseases 3 , 4 , 5 , 6 . Such regions, which were originally termed ‘gene deserts’ owing to their lack of coding genes, often contain GWAS hits but are poorly understood. To test for a shared disease mechanism, we performed co-localization analyses and confirmed that the genetic basis for every disease was the same, meaning that a common causal variant(s) and a shared molecular effect was responsible (Fig. 1a and Extended Data Fig. 1 ). As these heterogeneous diseases are all immune mediated, we reasoned that this locus must contain a distal enhancer that functioned in immune cells. By examining H3K27ac chromatin immunoprecipitation–sequencing (ChIP–seq) data, which marks active enhancers and promoters, we identified a monocyte/macrophage-specific enhancer within the locus (Fig. 1b ). Monocytes and macrophages have a key role in many immune-mediated diseases, producing cytokines that are often targeted therapeutically 14 .
a , Disease associations at chr21q22. The red points denote the IBD 99% credible set. Co-localization results for each disease versus IBD. PP.H3, posterior probability of independent causal variants; PP.H4, posterior probability of shared causal variant. b , Immune cell H3K27ac ChIP–seq at chr21q22. IBD GWAS results are shown. NK cells, natural killer cells. rpm, reads per million. c , The ETS2 eQTL in resting monocytes, with co-localization versus IBD association. Macrophage promoter-capture Hi-C (pcHi-C) data at the disease-associated locus. d , Experimental schematic for studying the chr21q22 locus in inflammatory (TPP) macrophages. e , ETS2 , BRWD1 and PSMG1 mRNA expression during TPP stimulation, measured using PrimeFlow RNA assays. Data are from one representative donor out of four. f , Relative ETS2 , BRWD1 and PSMG1 expression (mean fluorescence intensity (MFI)) in chr21q22-edited macrophages versus unedited cells. n = 4. Data are mean ± s.e.m. Statistical analysis was performed using two-way analysis of variance (ANOVA)). g , SuSiE fine-mapping posterior probabilities for IBD-associated SNPs at chr21q22 (99% credible set). h , Macrophage MPRA at chr21q22. Data are oligo coverage (top), enhancer activity (sliding-window analysis with significant enhancer activity highlighted; middle) and expression-modulating effects of SNPs within the enhancer (bottom). For the box plots, the centre line shows the median, the box limits show the interquartile range, and the whiskers represent the minimum and maximum values. n = 8. False-discovery rate (FDR)-adjusted P values were calculated using QuASAR-MPRA (two-sided). i , Inflammatory macrophage PU.1 ChIP–seq peaks at chr21q22. Bottom, magnification of the location of rs2836882 and the nearest predicted PU.1 motif. j , BaalChIP analysis of allele-specific PU.1 ChIP–seq binding at rs2836882 in two heterozygous macrophage datasets (data are mean ± 95% posterior distribution of allelic balance). Total counts shown as a pie chart. k , Allele-specific ATAC–seq reads at rs2836882 in monocytes from 16 heterozygous donors (including healthy controls and patients with ankylosing spondylitis). Statistical analysis was performed using two-sided Wilcoxon matched-pair tests. l , H3K27ac ChIP–seq data from risk (top) or non-risk (bottom) allele homozygotes at rs2836882. Data are shown from two out of four donors. FDR-corrected P values were calculated using MEDIPS (two-sided). The diagrams in d and e were created using BioRender.
Source Data
We next sought to identify the gene regulated by this enhancer. Although the associated locus lacks coding genes, there are several nearby candidates that have been highlighted in previous studies, including PSMG1 , BRWD1 and ETS2 (refs. 3 , 4 , 5 , 6 , 15 ) (Fig. 1a ). Using promoter-capture Hi-C and expression quantitative locus (eQTL) data from human monocytes ( Methods ), we found that the disease-associated locus physically interacts with the promoter of ETS2 —the most distant candidate gene (around 290 kb away)—and that the risk haplotype correlates with higher ETS2 expression (Fig. 1c ). Indeed, increased ETS2 expression in monocytes and macrophages, either at rest or after early exposure to bacteria, was found to have the same genetic basis as inflammatory disease risk (Extended Data Fig. 1c ). To directly confirm that ETS2 was causal, we used CRISPR–Cas9 to delete the 1.85 kb enhancer region in primary human monocytes before culturing these cells with inflammatory ligands, including TNF (a pro-inflammatory cytokine), prostaglandin E2 (a pro-inflammatory lipid) and Pam3CSK4 (a TLR1/2 agonist) (TPP model; Fig. 1d and Extended Data Fig. 2a–c ). This model was designed to mimic chronic inflammation 16 , and better resembles disease macrophages than classical IFNγ-driven or IL-4-driven models 17 (Extended Data Fig. 2 ). As flow cytometry antibodies were not available for the candidate genes, we used PrimeFlow to measure the dynamics of mRNA expression and detected increased levels of all three genes ( ETS2 , BRWD1 and PSMG1 ) after TPP stimulation of unedited monocytes (Fig. 1e ). Deletion of the chr21q22 enhancer did not affect BRWD1 or PSMG1 expression, but the upregulation of ETS2 was profoundly reduced (Fig. 1f ), confirming that this pleiotropic locus contains a distal ETS2 enhancer.
To identify the causal variant, we performed statistical fine-mapping in a large IBD GWAS 3 . Unfortunately, this did not resolve the association owing to high linkage disequilibrium between candidate single-nucleotide polymorphisms (SNPs) ( Methods and Fig. 1g ). We therefore used a functional approach to first delineate the active enhancers at the locus, and then assess whether any candidate SNPs might alter enhancer activity. This method, massively parallel reporter assay (MPRA), simultaneously tests enhancer activity in thousands of short DNA sequences by coupling each to a uniquely barcoded reporter gene 18 . Sequences that alter gene expression are identified by normalizing the barcode counts in mRNA, extracted from transfected cells, to their matching counts in the input DNA library. After adapting MPRA for primary macrophages ( Methods and Extended Data Fig. 3 ), we synthesized a pool of overlapping oligonucleotides to tile the 2 kb region containing all candidate SNPs, and included oligonucleotides with either risk or non-risk alleles for every variant. The resulting library was transfected into inflammatory macrophages from multiple donors, ensuring that a physiological repertoire of transcription factors could interact with the genomic sequences. Using a sliding-window analysis, we identified a single 442 bp focus of enhancer activity (chromosome 21: 40466236–40466677, hg19; Fig. 1h ) that contained three (out of seven) candidate SNPs. Two of these polymorphisms were transcriptionally inert, but the third (rs2836882) had the strongest expression-modulating effect of any candidate SNP, with the risk allele (G) increasing transcription, consistent with the ETS2 eQTL (Fig. 1h and Extended Data Fig. 1b ). This SNP was in the credible set of every co-localizing molecular trait, and lay within a macrophage PU.1 ChIP–seq peak (Fig. 1i ). PU.1 is a non-classical pioneer factor in myeloid cells 19 that can bind to DNA, initiate chromatin remodelling (thereby enabling other transcription factors to bind) and activate transcription 20 . To determine whether rs2836882 might affect PU.1 binding, we identified PU.1 ChIP–seq data from heterozygous macrophages and tested for allelic imbalances in binding. Despite not lying within a canonical PU.1 motif, strong allele-specific binding was detected, with over fourfold greater binding to the rs2836882 risk allele (Fig. 1i,j ). This was replicated by genotyping PU.1-bound DNA in macrophages from five heterozygous donors (Extended Data Fig. 4a–f ). Moreover, assay for transposase-accessible chromatin with sequencing (ATAC–seq) analysis of monocytes and macrophages from rs2836882 heterozygotes revealed allelic differences in chromatin accessibility that were consistent with differential binding of a pioneer factor (Fig. 1k and Extended Data Fig. 4g ).
To test for allele-specific enhancer activity at the endogenous locus, we performed H3K27ac ChIP–seq analysis of inflammatory macrophages from rs2836882 major and minor allele homozygotes. While most chr21q22 enhancer peaks were similar between these donors, the enhancer activity overlying rs2836882 was significantly stronger in major (risk) allele homozygotes (Fig. 1l and Extended Data Fig. 4h ), contributing to an approximate 2.5-fold increase in activity across the locus (Extended Data Fig. 4i ). Collectively, these data reveal a mechanism whereby the putative causal variant at chr21q22—identified by its functional effects in primary macrophages—promotes binding of a pioneer factor, enhances chromatin accessibility and increases activity of a distal ETS2 enhancer.
Macrophage inflammation requires ETS2
ETS2 is an ETS-family transcription factor and proto-oncogene 21 , but its exact role in human macrophages is unclear, with previous studies using either cell lines or complex mouse models and assessing a limited number of potential targets 22 , 23 , 24 , 25 , 26 . This has led to contradictory reports, with ETS2 being described as both necessary and redundant for macrophage development 27 , 28 , and both pro- and anti-inflammatory 22 , 23 , 24 , 25 , 26 . To clarify the role of ETS2 in human macrophages, and determine how dysregulated ETS2 expression might contribute to disease, we first used a CRISPR–Cas9-based loss-of-function approach (Fig. 2a ). To control for off-target effects, two gRNAs targeting different ETS2 exons were designed, validated and individually incorporated into Cas9 ribonucleoproteins for transfection into primary monocytes. These produced on-target editing in around 90% and 79% of cells, respectively, and effectively reduced ETS2 expression (Extended Data Fig. 2d–f ). Cell viability and macrophage marker expression were unaffected, suggesting that ETS2 was not required for macrophage survival or differentiation (Extended Data Fig. 2g,h ). By contrast, pro-inflammatory cytokine production, including IL-6, IL-8 and IL-1β, was markedly reduced after ETS2 disruption (Fig. 2b ), whereas IL-10—an anti-inflammatory cytokine—was less affected. TNF was not assessed as it had been added exogenously. We next investigated whether other macrophage functions were affected. Using fluorescently labelled particles that are detectable by flow cytometry, we found that phagocytosis was similarly impaired after ETS2 disruption (Fig. 2c ). We also tested extracellular reactive oxygen species (ROS) production—a major contributor to inflammatory tissue damage 29 . Disrupting ETS2 profoundly reduced the macrophage oxidative burst—most likely by decreasing expression of key NADPH oxidase components (Fig. 2d and Extended Data Fig. 5a ). Together, these data suggest that ETS2 is essential for multiple inflammatory functions in human macrophages.
a , Experimental schematic for studying ETS2 in inflammatory (TPP) macrophages. The diagram was created using BioRender. b , Cytokine secretion after ETS2 disruption. Heat map of relative cytokine levels from ETS2 -edited versus unedited macrophages. n = 8. c , Phagocytosis of fluorescently labelled zymosan particles by ETS2 -edited and unedited macrophages (non-targeting control (NTC)) (left). Data are from one representative donor out of seven. Right, the phagocytosis index (the product of the proportion and MFI of phagocytosing cells). n = 7. d , ROS production by ETS2 -edited and unedited macrophages. Data from one representative donor out of six (left). Right, NADPH oxidase component expression in ETS2 -edited and unedited macrophages (western blot densitometry). n = 7. Source gels are shown in Supplementary Fig. 1 . RLU, relative light units. e , RNA-seq analysis of differentially expressed genes in ETS2 -edited versus unedited TPP macrophages (limma with voom transformation, two-sided). n = 8. The horizontal line denotes the FDR-adjusted significance threshold. f , fGSEA of differentially expressed genes between ETS2 -edited and unedited TPP macrophages. The results of selected GO Biological Pathways are shown. The dot size denotes the unadjusted P value (two-sided), and the colour denotes normalized enrichment score (NES). g , The log 2 [fold change (FC)] of genes differentially expressed by chr21q22 enhancer deletion, plotted against their fold change after ETS2 editing. The percentages denote upregulated (red) and downregulated (blue) genes. The coloured points (blue or red) represent differentially expressed genes after ETS2 editing (FDR < 0.1, two-sided). For c and d , data are mean ± s.e.m. Statistical analysis was performed using two-sided Wilcoxon tests ( b – d ); * P < 0.05.
To understand the molecular basis for these effects, we performed RNA sequencing (RNA-seq) of ETS2 -edited and unedited inflammatory macrophages from multiple donors. Disrupting ETS2 led to widespread transcriptional changes, with reduced expression of many inflammatory genes (Fig. 2e ). These included cytokines (such as TNFSF10/TRAIL , TNFSF13 , IL1A and IL1B ), chemokines (such as CXCL3 , CXCL5 , CCL2 and CCL5 ), secreted effector molecules (such as S100A8 , S100A9 , MMP14 and MMP9 ), cell surface receptors (such as FCGR2A , FCGR2C and TREM1 ), pattern-recognition receptors (such as TLR2 , TLR6 and NOD2 ) and signalling molecules (such as MAP2K , GPR84 and NLRP3 ). To better characterize the pathways affected, we performed gene set enrichment analysis (fGSEA) using the Gene Ontology (GO) Biological Pathways dataset. This corroborated the functional deficits, with the most negatively enriched pathways (downregulated by ETS2 disruption) being related to macrophage activation, inflammatory cytokine production, phagocytosis and ROS production (Fig. 2f ). Genes involved in macrophage migration were also downregulated, but those relating to monocyte-to-macrophage differentiation were unaffected—consistent with ETS2 being required for inflammatory functions but not for monocyte-derived macrophage development. Fewer genes were upregulated after ETS2 disruption (Fig. 2e ), but positive enrichment was noted for aerobic respiration and oxidative phosphorylation (OXPHOS; Fig. 2f )—metabolic processes that are linked to anti-inflammatory phenotypes 30 . Notably, these transcriptional effects were not due to major changes in chromatin accessibility, although enhancer activity was generally reduced (Extended Data Fig. 2j,k ). As expected, deletion of the chr21q22 enhancer phenocopied both the transcriptional and functional effects of disrupting ETS2 (Fig. 2g and Extended Data Fig. 5a–e ). Collectively, these data identify an essential role for ETS2 in macrophage inflammatory responses, which could explain why dysregulated ETS2 expression predisposes to disease. Indeed, differential expression of ETS2-regulated genes was observed in resting (M0) macrophages from patients with IBD stratified by rs2836882 genotype (matched for age, sex, therapy and disease activity) (Extended Data Fig. 5f ).
ETS2 coordinates macrophage inflammation
We next studied the effects of increasing ETS2 expression, as this is what drives disease risk. To do this, we optimized a method for controlled overexpression of target genes in primary macrophages through transfection of in vitro transcribed mRNA that was modified to minimize immunogenicity (Fig. 3a , Methods and Extended Data Fig. 3f ). Resting, non-activated macrophages were transfected with ETS2 mRNA or its reverse complement, thereby controlling for mRNA quantity, length and purine/pyrimidine composition (Fig. 3b ). After transfection, cells were exposed to low-dose lipopolysaccharide to initiate a low-grade inflammatory response that could potentially be amplified (Fig. 3a ). We found that overexpressing ETS2 increased pro-inflammatory cytokine secretion, while IL-10 was again less affected (Extended Data Fig. 3g ). To better characterize this response, we performed RNA-seq and re-examined the inflammatory pathways that required ETS2 . Notably, all of these pathways—including macrophage activation, cytokine production, ROS production, phagocytosis and migration—were induced in a dose-dependent manner by ETS2 overexpression, with greater enrichment of every pathway when more ETS2 mRNA was transfected (Fig. 3c ). This shows that ETS2 is both necessary and sufficient for inflammatory responses in human macrophages, consistent with being a central regulator of effector functions, with dysregulation directly linked to disease.
a , Experimental schematic for studying the effects of ETS2 overexpression. The diagram was created using BioRender. b , ETS2 mRNA levels in transfected ( n = 8) or untransfected (from a separate experiment) macrophages. Data are mean ± s.e.m. CPM, counts per million. c , fGSEA analysis of differentially expressed genes between ETS2 -overexpressing and control macrophages. Results shown for pathways downregulated by ETS2 disruption. The dot size denotes the unadjusted P value (two-sided), the colour denotes NES and the border colour denotes the quantity of transfected mRNA. d , fGSEA analysis of a Crohn’s disease intestinal macrophage signature in ETS2 -overexpressing macrophages (versus control). FDR P -value, two-sided (top). Heat map of the relative expression of leading-edge genes after ETS2 overexpression (500 ng mRNA; bottom). e , Enrichment of macrophage signatures from patients with the indicated diseases in ETS2 -overexpressing macrophages (versus control). The colour denotes the disease category, the numbers denote the NES and the dashed line denotes FDR = 0.05. The Crohn’s disease signature is from a different study to that shown in d . AS, ankylosing spondylitis. f , SNPsea analysis of genes tagged by 241 IBD SNPs within ETS2 -regulated genes (red) and known IBD pathways (black). Significant pathways (Bonferroni-corrected P < 0.05) are indicated by hash symbols (#).
ETS2 has a key pathogenic role in IBD
To test whether ETS2 contributes to macrophage phenotypes in disease, we compared the effects of overexpressing ETS2 in resting macrophages with a single-cell RNA-seq (scRNA-seq) signature from intestinal macrophages in Crohn’s disease 31 . ETS2 overexpression induced a transcriptional state that closely resembled disease macrophages, with core (leading edge) enrichment of most signature genes, including several therapeutic targets (Fig. 3d ). Similar enrichment was observed with myeloid signatures from other chr21q22-associated diseases and, to a lesser extent, from active bacterial infection, but not for signatures from influenza and tumour macrophages, suggesting that ETS2 was not simply inducing generic activation (Fig. 3e ).
Given the central role of ETS2 in inflammatory macrophages and the importance of these cells in disease, we hypothesized that other genetic associations would also implicate this pathway. A major goal of GWAS was to identify disease pathways, but this has proven to be challenging due to a paucity of confidently identified causal genes and variants 9 . To determine whether the macrophage ETS2 pathway was enriched for disease genetics, we focused on IBD as this has more GWAS hits than any other chr21q22-associated disease. Encouragingly, a network of 33 IBD-associated genes in intestinal mucosa was previously found to be enriched for predicted ETS2 motifs 32 . Examining the genes that were consistently downregulated in ETS2 -edited macrophages (adjusted P ( P adj ) < 0.05 for both gRNAs), we identified over 20 IBD-risk-associated genes, including many thought to be causal at their respective loci 3 , 33 (Extended Data Table 1 ). These included genes that are known to affect macrophage biology (such as SP140 , LACC1 , CCL2 , CARD9 , CXCL5 , TLR4 , SLAMF8 and FCGR2A ) and some that are highly expressed in macrophages but not linked to specific pathways (such as ADCY7 , PTPRC , TAGAP , PTAFR and PDLIM5 ). A polygenic risk score comprising these variants associated with features of more severe IBD across 18,249 patients, including earlier disease onset, increased the need for surgery, and stricturing or fistulating complications in Crohn’s disease (Extended Data Fig. 6a–h ). To better test the enrichment of IBD GWAS hits in ETS2-mediated inflammation, and compare this with known disease pathways, we used SNPsea 34 —a method to identify pathways affected by disease loci. In total, 241 IBD loci were tested for enrichment in 7,658 GO Biological Pathways and 2 overlapping lists of ETS2-regulated genes (either those downregulated by ETS2 disruption or upregulated by ETS2 overexpression). Statistical significance was computed using 5 million matched null SNP sets, and pathways implicated by IBD genetics were extracted for comparison. Notably, IBD-associated SNPs were more significantly enriched in the macrophage ETS2 pathway than in many IBD pathways, with not a single null SNP set being more enriched in either ETS2-regulated gene list (Fig. 3f and Extended Data Fig. 6i ). SNPs associated with primary sclerosing cholangitis (PSC), ankylosing spondylitis and Takayasu’s arteritis were also enriched in ETS2-target genes (Extended Data Fig. 6j ). Collectively, this suggests that macrophage ETS2 signalling has a central role in multiple inflammatory diseases.
ETS2 has distinct inflammatory effects
We next investigated how ETS2 might control such diverse macrophage functions. Studying ETS2 biology is challenging because no ChIP-grade antibodies exist, precluding direct identification of its transcriptional targets. We therefore first used a guilt-by-association approach to identify genes that were co-expressed with ETS2 across 67 human macrophage activation conditions (comprising 28 stimuli and various durations of exposure) 16 . This identified PFKFB3 —encoding the rate-limiting enzyme of glycolysis—as the most highly co-expressed gene, with HIF1A also highly co-expressed (Fig. 4a ). Together, these genes facilitate a ‘glycolytic switch’ that is required for myeloid inflammatory responses 35 . We therefore hypothesized that ETS2 might control inflammation through metabolic reprogramming—a possibility supported by OXPHOS genes being negatively correlated with ETS2 (Fig. 4a ) and upregulated after ETS2 disruption (Fig. 2f ). To assess the metabolic consequences of disrupting ETS2 , we quantified label incorporation from 13 C-glucose in edited and unedited TPP macrophages using gas chromatography coupled with mass spectrometry (GC–MS). Widespread modest reductions in labelled and total glucose metabolites were detected after ETS2 disruption (Fig. 4b and Extended Data Fig. 7a–c ). This affected both glycolytic and tricarboxylic acid (TCA) cycle metabolites, with significant reductions in lactate, a hallmark of anaerobic glycolysis, and succinate, a key inflammatory metabolite 36 . These results are consistent with glycolytic suppression, with reductions in TCA metabolites being due to reduced flux into TCA and increased consumption by mitochondrial OXPHOS 37 . To determine whether metabolic changes accounted for ETS2-mediated inflammatory effects, we treated ETS2 -edited macrophages with roxadustat—a HIF1α stabilizer that promotes glycolysis. This had the predicted effect on glycolysis and OXPHOS genes, but did not rescue the effects of ETS2 disruption, either transcriptionally or functionally (Fig. 4c and Extended Data Fig. 7d,e ). Thus, while disrupting ETS2 impairs macrophage glycometabolism, this does not fully explain the differences in inflammation.
a , Genes co-expressed with ETS2 across 67 monocyte/macrophage activation conditions. The dotted lines denote FDR-adjusted P < 0.05. b , The effect of ETS2 disruption on glucose metabolism. The colour denotes median log 2 -transformed fold change in label incorporation from 13 C-glucose in ETS2 -edited versus unedited cells. The bold black border denotes P < 0.05 (Wilcoxon matched-pairs, two-sided). n = 6. Sec., secreted. c , fGSEA analysis of differentially expressed genes between ETS2 -edited and unedited macrophages that were treated with roxadustat or vehicle. Results shown for pathways downregulated by ETS2 disruption. d , Enrichment heat maps of macrophage ETS2 CUT&RUN peaks (IDR cut-off 0.01, n = 2) in 4 kb peak-centred regions from ATAC–seq (accessible chromatin), H3K4me3 ChIP–seq (active promoters) and H3K27ac ChIP–seq (active regulatory elements). e , Functional annotations of ETS2-binding sites (using gene coordinates and TPP macrophage H3K27ac ChIP–seq data). f , ETS2 motif enrichment in CUT&RUN peaks (hypergeometric P value, two-sided). g , ETS2 binding, chromatin accessibility (ATAC–seq) and regulatory activity (H3K27ac) at selected loci. h , Intersections between genes with ETS2 peaks in their core promoters or cis -regulatory elements and genes upregulated (Up) or downregulated (Dn) after ETS2 editing (KO) or overexpression (OE). The vertical bars denote the size of overlap for lists indicated by connected dots in the bottom panel. The horizontal bars denote the percentage of gene list within intersections. i , ETS2 binding, PU.1 binding, chromatin accessibility and enhancer activity at chr21q22. Predicted ETS2-binding sites (red) and PU.1-binding sites (purple) shown below. The dashed line is positioned at rs2836882.
We therefore revisited whether we could directly identify ETS2-target genes. As ChIP–seq involves steps that can alter protein epitopes and prevent antibody binding (such as fixation) we tested whether any anti-ETS2 antibodies might work for cleavage under targets and release using nuclease (CUT&RUN), which does not require these steps. One antibody identified multiple significantly enriched genomic regions (peaks), of which 6,560 were reproducibly detected across two biological replicates with acceptable quality metrics 38 (Fig. 4d ). These peaks were mostly located in active regulatory regions (90% in promoters or enhancers; Fig. 4d,e ) and were highly enriched for both a canonical ETS2 motif (4.02-fold versus global controls; Fig. 4f ) and for motifs of known ETS2 interactors, including FOS, JUN and NF-κB 39 (Extended Data Fig. 7f ). After combining the biological replicates to improve peak detection, we identified ETS2 binding at genes involved in multiple inflammatory functions, including NCF4 (ROS production), NLRP3 (inflammasome activation) and TLR4 (bacterial pattern recognition) (Fig. 4g ). Overall, 48.3% (754 out of 1,560) of genes dysregulated after ETS2 disruption and 50.3% (1,078 out of 2,153) of genes dysregulated after ETS2 overexpression contained an ETS2-binding peak within their core promoter or cis -regulatory elements (Fig. 4h ). Notably, ETS2 targets included HIF1A , PFKFB3 and other glycolytic genes (such as GPI , HK2 and HK3 ), consistent with the observed metabolic changes being directly induced as part of this complex inflammatory programme. Notably, we also detected ETS2 binding at the chr21q22 enhancer (Fig. 4i ). This is consistent with reports that PU.1 and ETS2 can interact synergistically 40 , and suggests that ETS2 might contribute to the activity of its own enhancer. Indeed, manipulating ETS2 expression altered enhancer activity in a manner consistent with positive autoregulation (Extended Data Fig. 7g–i ). Together, these data implicate ETS2 as a central regulator of monocyte and macrophage inflammatory responses that is able to direct a multifaceted effector programme and create a metabolic environment that is permissive for inflammation.
Targeting the ETS2 pathway in disease
To assess how ETS2 affects macrophage heterogeneity in diseased tissue, and whether this could be targeted therapeutically, we examined intestinal scRNA-seq data from patients with Crohn’s disease and healthy control individuals 41 . Within myeloid cells, seven clusters were detected and identified using established markers and/or previous literature (Fig. 5a,b ). Inflammatory macrophages (cluster 1, expressing CD209, CCL4, IL1B and FCGR3A) and inflammatory monocytes (cluster 2, expressing S100A8/A9, TREM1, CD14 and MMP9) were expanded in disease, as previously described 42 , and expressed ETS2 and ETS2-regulated genes more highly than other clusters, including tissue-resident macrophages (cluster 0, expressing C1QA, C1QB, FTL and CD63) and conventional dendritic cells (cluster 5, expressing CLEC9A, CADM1 and XCR1) (Fig. 5a,b and Extended Data Fig. 8a ). Using spatial transcriptomics, a similar increase in inflammatory macrophages was observed in PSC liver tissue, with these cells being closely apposed to cholangiocytes—the main target of pathology (Fig. 5c–e ). Notably, expression of ETS2-regulated genes was higher the closer macrophages were to cholangiocytes (Fig. 5f and Extended Data Fig. 8b ). Indeed, using bulk RNA-seq data, we found that the transcriptional footprint of ETS2 was detectable in affected tissues from multiple chr21q22-associated diseases (Extended Data Fig. 8c ).
a , Myeloid cell clusters in intestinal scRNA-seq from Crohn’s disease and health (top). Middle, scaled expression of ETS2-regulated genes (downregulated by ETS2 disruption). Bottom, the source of cells (disease or health). b , Scaled expression of selected genes. c , Spatial transcriptomics of PSC and healthy liver. n = 4. The images show representative fields of view (0.51 mm × 0.51 mm) with cell segmentation and semisupervised clustering. The main key (left and middle below images) denotes InSituType cell types; clusters a–e (far right key) are unannotated cell populations. Hep., hepatocyte; LSECs, liver sinusoidal endothelial cells; non-inflamm. macs, non-inflammatory macrophages. d , The number of macrophages within the indicated distances of cholangiocytes. e , The distance from cholangiocytes to the nearest macrophage. Data are shown as Tukey box and whisker plots. Statistical analysis was performed using two-tailed Mann–Whitney U -tests. Data in d and e are from 10,532 PSC and 13,322 control cholangiocytes. f , Scaled expression of ETS2-regulated genes in 21,067 PSC macrophages at defined distances from cholangiocytes (excluding genes used to define macrophage subsets). g , Classes of drugs that phenocopy ETS2 disruption (from the NIH LINCS database). h , fGSEA results for NIH LINCS drug signatures. Significant MEK inhibitor signatures are coloured by molecule. i , The log 2 [fold change] of differentially expressed genes after chr21q22 enhancer deletion, plotted against their fold change after MEK inhibition. The percentages indicate the proportion of upregulated (red) and downregulated genes (blue). The coloured points (blue or red) were differentially expressed after MEK inhibition (FDR < 0.1). j , fGSEA of differentially expressed genes between MEK-inhibitor-treated and control TPP macrophages. Results are shown for pathways downregulated by ETS2 disruption. The dot size denotes the unadjusted P value (two-sided) and the colour denotes the NES. k , IBD biopsy cytokine release with PD-0325901, infliximab or vehicle control. l , GSVA enrichment scores for chr21q22-downregulated genes in IBD biopsies after MEK inhibition. m , GSVA enrichment scores of a biopsy-derived molecular inflammation score (bMIS). Data are mean ± 95% CI ( f and l ) and mean ± s.e.m. ( k and m ). Statistical analysis was performed using two-sided paired t -tests. n = 10 ( k ), n = 9 ( l ). ** P < 0.01, *** P < 0.001, **** P < 0.0001.
We next examined whether this pathway could be targeted pharmacologically. Specific ETS2 inhibitors do not exist and structural analyses indicate that there is no obvious allosteric inhibitory mechanism 43 . We therefore used the NIH LINCS database to identify drugs that might modulate ETS2 activity 7 . This contains over 30,000 differentially expressed gene lists from cell lines exposed to around 6,000 small molecules. Using fGSEA, 906 signatures mimicked the effect of disrupting ETS2 ( P adj < 0.05), including several approved IBD therapies. The largest class of drugs was MEK inhibitors (Fig. 5g ), which are licensed for non-inflammatory human diseases (such as neurofibromatosis). This result was not due to a single compound, but rather a class effect with multiple MEK1/2 inhibitors downregulating ETS2-target genes (Fig. 5h ). This made biological sense, as MEK1/2, together with several other targets identified, are known regulators of ETS-family transcription factors (Fig. 5g ). Some of these compounds have shown benefit in animal colitis models 44 , although this is often a poor indicator of clinical efficacy, as several IBD treatments are ineffective in mice and many compounds that improve mouse models are ineffective in humans 45 . To test whether MEK inhibition abrogates ETS2-driven inflammation in human macrophages, we treated TPP macrophages with PD-0325901, a selective non-ATP competitive MEK inhibitor. Potent anti-inflammatory activity was observed that phenocopied the effects of disrupting ETS2 or the chr21q22 enhancer (Fig. 5i,j and Extended Data Fig. 9a–c ). To further assess the therapeutic potential, we cultured intestinal biopsies from active, untreated IBD with either a MEK inhibitor or a negative or positive control ( Methods ). MEK inhibition reduced inflammatory cytokine release to similar levels as infliximab (an anti-TNF antibody that is widely used for IBD; Fig. 5k ). Moreover, ETS2-regulated gene expression was reduced (Fig. 5l and Extended Data Fig. 9d ) and there was improvement in a transcriptional inflammation score 46 (Fig. 5m ). Together, these data show that targeting an upstream regulator of ETS2 can abrogate pathological inflammation in a chr21q22-associated disease, and may be useful therapeutically.
Arguably the greatest challenge in modern genetics is to translate the success of GWAS into a better understanding of disease. Here, by studying a pleiotropic disease locus, we identify a central regulator of human macrophage inflammation and a pathogenic pathway that is potentially druggable. These findings also provide clues to the gene–environment interactions at this locus, highlighting a potential role for ETS2 in macrophage responses to bacteria. This would provide a balancing selection pressure that might explain why the risk allele remains so common (frequency of around 75% in Europeans and >90% in Africans) despite first being detected in archaic humans over 500,000 years ago (Extended Data Fig. 10 ).
Although ETS2 was reported to have pro-inflammatory effects on individual genes 24 , 25 , the full extent of its inflammatory programme—with effects on ROS production, phagocytosis, glycometabolism and macrophage activation—was unclear. Moreover, without direct proof of ETS2 targets, nor studies in primary human cells, it was difficult to reconcile reports of anti-inflammatory effects at other genes 23 , 26 . By systematically characterizing the effects of ETS2 disruption and overexpression in human macrophages, we identify an essential role in inflammation, delineate the mechanisms involved and show how ETS2 can induce pathogenic macrophage phenotypes. Increased ETS2 expression may also contribute to other human pathology. For example, Down’s syndrome (trisomy 21) was recently described as a cytokinopathy 47 , with basal increases in multiple inflammatory cytokines, including several ETS2 targets (such as IL-1β, TNF and IL-6). Whether the additional copy of ETS2 contributes to this phenotype is unknown, but warrants further study.
Blocking individual cytokines is a common treatment strategy in inflammatory disease 14 , but emerging evidence suggests that targeting several cytokines at once may be a better approach 48 . Blocking ETS2 signalling through MEK1/2 inhibition affects multiple cytokines, including TNF and IL-23, which are targets of existing therapies, and IL-1β, which is linked to treatment resistance 49 and not directly modulated by other small molecules (such as JAK inhibitors). However, long-term MEK inhibitor use may not be ideal owing to the physiological roles of MEK in other tissues, with multiple side-effects having been reported 50 . Targeting ETS2 directly—for example, through PROTACs—or selectively delivering MEK inhibitors to macrophages through antibody–drug conjugates could overcome this toxicity, and provide a safer means of blocking ETS2-driven inflammation.
In summary, using an intergenic GWAS hit as a starting point, we have identified a druggable pathway that is both necessary and sufficient for human macrophage inflammation. Moreover, we show how genetic dysregulation of this pathway—through perturbation of pioneer factor binding at a critical long-range enhancer—predisposes to multiple diseases. This highlights the considerable, yet largely untapped, opportunity to resolve disease biology from non-coding genetic associations.
Analysis of existing data relating to chr21q22
IBD GWAS summary statistics 3 were used to perform multiple causal variant fine-mapping using susieR 51 , with reference minor allele and LD information calculated from 503 European samples from 1000 Genomes phase 3 (ref. 52 ). All R analyses used v.4.2.1. Palindromic SNPs (A/T or C/G) and any SNPs that did not match by position or alleles were pruned before imputation using the ssimp equations reimplemented in R. This did not affect any candidate SNP at chr21q22. SuSiE fine-mapping results were obtained for ETS2 (identifier ENSG00000157557 or ILMN_1720158) in monocyte/macrophage datasets from the eQTL Catalogue 53 . Co-localization analyses were performed comparing the chr21q22 IBD association with summary statistics from other chr21q22-associated diseases 3 , 4 , 5 , 6 and monocyte/macrophage eQTLs 54 , 55 , 56 , 57 , 58 to determine whether there was a shared genetic basis for these different associations. This was performed using coloc (v.5.2.0) 59 using a posterior probability of H4 (PP.H4.abf) > 0.5 to call co-localization.
Raw H3K27ac ChIP–seq data from primary human immune cells were downloaded from Gene Expression Omnibus (GEO series GSE18927 and GSE96014 ) and processed as described previously 60 (code provided in the ‘Code availability’ section).
Processed promoter-capture Hi-C data 61 from 17 primary immune cell types were downloaded from OSF ( https://osf.io/u8tzp ) and cell type CHiCAGO scores for chr21q22-interacting regions were extracted.
Monocyte-derived macrophage differentiation
Leukocyte cones from healthy donors were obtained from NHS Blood and Transplant (Cambridge Blood Donor Centre, Colindale Blood Centre or Tooting Blood Donor Centre). Peripheral blood mononuclear cells (PBMCs) were isolated by density centrifugation (Histopaque 1077, Sigma-Aldrich) and monocytes were positively selected using CD14 Microbeads (Miltenyi Biotec). Macrophage differentiation was performed either using conditions that model chronic inflammation (TPP) 16 : 3 days GM-CSF (50 ng ml −1 , Peprotech) followed by 3 days GM-CSF, TNF (50 ng ml −1 , Peprotech), PGE 2 (1 μg ml −1 , Sigma-Aldrich) and Pam 3 CSK4 (1 μg ml −1 , Invivogen); or, to produce resting (M0) macrophages: 6 days M-CSF (50 ng ml −1 , Peprotech). All cultures were performed at 37 °C under 5% CO 2 in antibiotic-free RPMI1640 medium containing 10% FBS, GlutaMax and MEM non-essential amino acids (all Thermo Fisher Scientific). Cells were detached using Accutase (BioLegend).
Identifying a model of chronic inflammatory macrophages
Human monocyte/macrophage gene expression data files ( n = 314) relating to 28 different stimuli with multiple durations of exposure (collectively comprising 67 different activation conditions) were downloaded from the GEO ( GSE47189 ) and quantile normalized. Data from biological replicates were summarized to the median value for every gene. Gene set variation analysis 62 (using the GSVA package in R) was performed to identify the activation condition that most closely resembled CD14 + monocytes/macrophages from active IBD using disease-associated lists of differentially expressed genes 63 .
CRISPR–Cas9 editing of primary human monocytes
gRNA sequences were designed using CRISPick and synthesized by IDT (Supplementary Table 3 ). Alt-R CRISPR–Cas9 negative control crRNA 1 (IDT) was used as a non-targeting control. Cas9–gRNA ribonucleoproteins were assembled as described previously 60 and nucleofected into 5 × 10 6 monocytes in 100 μl nucleofection buffer (Human Monocyte Nucleofection Kit, Lonza) using a Nucleofector 2b (Lonza, program Y-001). After nucleofection, monocytes were immediately transferred into 5 ml of prewarmed culture medium in a six-well plate, and differentiated into macrophages under TPP conditions. The editing efficiency was quantified by PCR amplification of the target region in extracted DNA. All primer sequences are provided in Supplementary Table 3 . The editing efficiency at the chr21q22 locus was measured by quantification of amplified fragments (2100 Bioanalyzer, Agilent) as previously described 60 . The editing efficiency for individual gRNAs was assessed using the Inference of CRISPR Edits tool 64 (ICE, Synthego).
PrimeFlow RNA assay
RNA abundance was quantified by PrimeFlow (Thermo Fisher Scientific) in chr21q22-edited and unedited (NTC) cells on days 0, 3, 4, 5 and 6 of TPP differentiation. Target probes specific for ETS2 (Alexa Fluor 647), BRWD1 (Alexa Fluor 568) and PSMG1 (Alexa Fluor 568) were used according to the manufacturer’s instructions. Data were collected using FACS Diva software and analysed using FlowJo v10 (BD Biosciences).
Overlapping oligonucleotides containing 114 nucleotides of genomic sequence were designed to tile the region containing chr21q22 candidate SNPs (99% credible set) at 50 bp intervals. Six technical replicates were designed for every genomic sequence, each tagged by a unique 11-nucleotide barcode. Additional oligonucleotides were included to test the expression-modulating effect of every candidate SNP in the 99% credible set. Allelic constructs were designed as described previously 60 and tagged by 30 unique 11-nucleotide barcodes. Positive and negative controls were included as described previously 60 . 170-nucleotide oligonucleotides were synthesized as part of a larger MPRA pool (Twist Biosciences) containing the 16-nucleotide universal primer site ACTGGCCGCTTCACTG, 114-nucleotide variable genomic sequence, KpnI and XbaI restriction sites (TGGACCTCTAGA), an 11-nucleotide barcode and the 17-nucleotide universal primer site AGATCGGAAGAGCGTCG. Cloning into the MPRA vector was performed as described previously 60 . A suitable promoter for the MPRA vector (RSV) was identified by testing promoter activities in TPP macrophages. The MPRA vector library was nucleofected into TPP macrophages (5 µg vector into 5 × 10 6 cells) in 100 μl nucleofection buffer (Human Macrophage Nucleofection Kit, Lonza) using a Nucleofector 2b (program Y-011). To ensure adequate barcode representation, a minimum of 2 × 10 7 cells was nucleofected for every donor ( n = 8). After 24 h, RNA was extracted and sequencing libraries were made from mRNA or DNA input vector as described previously 60 . Libraries were sequenced on the Illumina HiSeq2500 high-output flow-cell (50 bp, single-end reads). Data were demultiplexed and converted to FASTQ files using bcl2fastq and preprocessed as previously described using FastQC 60 . To identify regions of enhancer activity, a paired t -test was first performed to identify genomic sequences that enhanced transcription and a sliding-window analysis (300 bp window) was then performed using the les package in R. Expression-modulating variants were identified using QuASAR-MPRA 65 , as described previously 60 .
Publicly available PU.1 ChIP–seq datasets from human macrophages were downloaded from GEO, and BAM files were examined (IGV genome browser) to identify heterozygous samples (that is, files containing both A and G allele reads at chr21:40466570; hg19). Two suitable samples were identified ( GSM1681423 and GSM1681429 ) and used for a Bayesian analysis of allelic imbalances in PU.1 binding (implemented in the BaalChIP package 66 in R) with correction for biases introduced by overdispersion and biases towards the reference allele.
Allele-specific PU.1 ChIP genotyping
A 100 ml blood sample was taken from five healthy rs2836882 heterozygotes (assessed by Taqman genotyping; Thermo Fisher Scientific). All of the participants provided written informed consent. Ethical approval was provided by the London–Brent Regional Ethics Committee (21/LO/0682). Monocytes were isolated from PBMCs using CD14 Microbeads (Miltenyi Biotec) and differentiated into inflammatory macrophages using TPP conditions 16 . After differentiation, macrophages were detached and cross-linked for 10 min in fresh medium containing 1% formaldehyde. Cross-linking was quenched with glycine (final concentration 0.125 M, 5 min). Nucleus preparation and shearing were performed as described previously 60 with 10 cycles sonication (30 s on/30 s off, Bioruptor Pico, Diagenode). PU.1 was immunoprecipitated overnight at 4 °C using a polyclonal anti-PU.1 antibody (1:25; Cell Signaling) using the SimpleChIP Plus kit (Cell Signaling). The ratio of rs2836882 alleles in the PU.1-bound DNA was quantified in duplicate by TaqMan genotyping (assay C 2601507_20). A standard curve was generated using fixed ratios of geneblocks containing either the risk or non-risk allele (200-nucleotide genomic sequence centred on rs2836882; Genewiz).
PU.1 MPRA ChIP–seq
The MPRA vector library was transfected into TPP macrophages from six healthy donors. Assessment of PU.1 binding to SNP alleles was performed as described previously 60 , with minimal sonication (to remove contaminants without chromatin shearing). Immunoprecipitation was performed as described above. Sequencing libraries were prepared as for MPRA and sequenced on the MiSeq system (50 bp, single-end reads).
ATAC–seq analysis
ATAC–seq in ETS2 -edited and unedited TPP macrophages was performed using the Omni-ATAC protocol 67 with the following modifications: the cell number was increased to 75,000 cells; the cell lysis time was increased to 5 min; the volume of Tn5 transposase in the transposition mixture was doubled; and the duration of the transposition step was extended to 40 min. Amplified libraries were cleaned using AMPure XP beads (Beckman Coulter) and sequenced on the NovaSeq6000 system (100 bp paired-end reads). Data were processed as described previously 68 . Differential ATAC–seq analysis was performed as described previously using edgeR and TMM normalization 69 . Allele-specific ATAC–seq analysis was performed in 16 heterozygous monocyte datasets from healthy controls and patients with ankylosing spondylitis 70 and in 2 deeply sequenced heterozygous TPP macrophage samples. For these analyses, sequencing reads at rs2836882 were extracted from preprocessed data using splitSNP ( https://github.com/astatham/splitSNP ) (see the ‘Code availability’ section).
H3K27ac ChIP–seq
H3K27ac ChIP–seq was performed as described previously 60 using an anti-H3K27ac antibody (1:250, Abcam) or an isotype control (1:500, rabbit IgG, Abcam). Sequencing libraries from TPP macrophages from major and minor allele homozygotes at rs2836882 (identified through the NIHR BioResource, n = 4) were sequenced on the HiSeq4000 system (50 bp, single-end reads). Sequencing libraries from ETS2 -edited and unedited TPP macrophages ( n = 3) or resting M0 macrophages overexpressing ETS2 or control mRNA ( n = 3) were sequenced on the NovaSeq6000 system (100 bp, paired-end reads). Raw data were processed, quality controlled and analysed as described previously using the Burrows-Wheeler Aligner 60 . Unpaired differential ChIP–seq analysis, to compare rs2836882 genotypes, was performed using MEDIPS 71 by dividing the 560 kb region around rs2836882 (chr21:40150000–40710000, hg19) into 5 kb bins. Paired differential ChIP–seq analyses, to assess the effect of perturbing ETS2 expression on enhancer activity, were performed using edgeR with TMM normalization 69 , 72 (with donor as covariate). Genome-wide analyses used consensus MACS2 peaks. Superenhancer activity was evaluated using Rank-Ordering of Super-Enhancers (ROSE). Chr21q22-based analyses used the enhancer coordinates that exhibited allele-specific activity (chr21:40465000–40470000, hg19). Code is provided for all data analysis (see the ‘Code availability’ section).
Assays of macrophage effector functions
Flow cytometry.
Expression of myeloid markers was assessed using flow cytometry (BD LSRFortessa X-20) with the following panel: CD11b PE/Dazzle 594 (BioLegend), CD14 evolve605 (Thermo Fisher Scientific), CD16 PerCP (BioLegend), CD68 FITC (BioLegend), Live/Dead Fixable Aqua Dead Cell Stain (Thermo Fisher Scientific) and Fc Receptor Blocking Reagent (Miltenyi). All antibodies were used at a dilution of 1:40; Live/Dead stained was used at 1:400 dilution. Data were collected using FACS Diva and analysed using FlowJo v.10 (BD Biosciences).
Cytokine quantification
Supernatants were collected on day 6 of TPP macrophage culture and frozen. Cytokine concentrations were quantified in duplicate by electrochemiluminescence using assays (Meso Scale Diagnostics, DISCOVERY WORKBENCH v.4.0).
Phagocytosis
Phagocytosis was assessed using fluorescently labelled Zymosan particles (Green Zymosan, Abcam) according to the manufacturer’s instructions. Cells were seeded at 10 5 cells per well in 96-well round-bottom plates. Cytochalasin D (10 μg ml −1 , Thermo Fisher Scientific) was used as a negative control. Phagocytosis was quantified by flow cytometry, and a phagocytosis index was calculated (the proportion of positive cells multiplied by their mean fluorescence intensity).
Extracellular ROS production
Extracellular ROS production was quantified using the Diogenes Enhanced Superoxide Detection Kit (National Diagnostics) according to the manufacturer’s protocol. Cells were seeded at a density of 10 5 cells per well and prestimulated with PMA (200 ng ml −1 , Sigma-Aldrich).
Western blotting
Western blotting was performed as described previously 73 using the following primary antibodies: mouse anti-gp91phox (1:2,000), mouse anti-p22phox (1:500; both Santa Cruz), rabbit anti-C17ORF62/EROS (1:1,000; Atlas), mouse anti-vinculin (Sigma-Aldrich). Loading controls were run on the same gel. Secondary antibodies were as follows: goat anti-rabbit IgG-horseradish or goat anti-mouse IgG-horseradish peroxidase (both 1:10,000; Jackson Immuno). Chemiluminescence was recorded on the ChemiDoc Touch imager (Bio-Rad) after incubation of the membrane with ECL (Thermo Fisher Scientific) or SuperSignal West Pico PLUS (Thermo Fisher Scientific) reagent. Densitometry analysis was performed using ImageJ.
RNA-seq analysis
RNA was isolated from macrophage lysates (AllPrep DNA/RNA Micro Kit, Qiagen) and sequencing libraries were prepared from 10 ng RNA using the SMARTer Stranded Total RNA-Seq Kit v2 Pico Input Mammalian (Takara) according to the manufacturer’s instructions. Libraries were sequenced on either the NextSeq 2000 (50 bp paired-end reads: CRISPR, roxadustat and PD-0325901 experiments) or NovaSeq 6000 (100 bp paired-end reads: overexpression experiments) system and preprocessed using MultiQC. Reads were trimmed using Trim Galore (Phred score 24) and filtered to remove reads <20 bp. Ribosomal reads (mapping to human ribosomal DNA complete repeating unit; GenBank: U13369 .1 ) were removed using BBSplit ( https://sourceforge.net/projects/bbmap/ ). Reads were aligned to the human genome (hg38) using HISAT2 (ref. 74 ) and converted to BAM files, sorted and indexed using SAMtools 75 . Gene read counts were obtained using the featureCounts program 76 from Rsubread using the GTF annotation file for GRCh38 (v.102). Differential expression analysis was performed in R using limma 77 with voom transformation and including donor as a covariate. Differential expression results are shown in Supplementary Tables 1 and 2 .
GSEA was performed using fGSEA 78 in R with differentially expressed gene lists ranked by t -statistic. Gene sets were obtained from GO Biological Pathways (MSigDB), experimentally derived based on differential expression analysis or sourced from published literature 31 , 42 , 70 , 79 , 80 , 81 , 82 , 83 , 84 , 85 , 86 . Specific details of disease macrophage signatures (Fig. 3f ) are provided as source data. GO pathways shown in Figs. 2 – 5 are as follows: GO:0002274, GO:0042116, GO:0097529, GO:0006909, GO:0071706, GO:0032732, GO:0032755, GO:0032757, GO:2000379, GO:0009060, GO:0006119 and GO:0045649. Statistical significance was calculated using the adaptive multilevel split Monte Carlo method.
IBD BioResource recall-by-genotype study
IBD patients who were rs2836882 major or minor allele homozygotes ( n = 11 of each) were identified through the NIHR IBD BioResource. Patients were matched for age, sex, treatment and disease activity, and all provided written informed consent. Ethical approval was provided by the London–Brent Regional Ethics Committee (21/LO/0682). A 50 ml blood sample was taken from all patients and M0 monocyte-derived macrophages were generated as described. After 6 days, cells were collected, lysed and RNA was extracted. Quantitative PCR analysis of a panel of ETS2-regulated genes was performed in triplicate after reverse transcription (SuperScript IV VILO, Thermo Fisher Scientific) using the Quantifast SYBR Green PCR kit (Qiagen) on the Roche LightCycler 480. Primer sequences are provided in Supplementary Table 3 and PPIA and RPLP0 were used as housekeeping genes. Expression values for each gene ( \({2}^{\Delta {c}_{T}}\) ) were scaled to a minimum 0 and maximum 1 to enable intergene comparison.
In vitro transcription
The cDNA sequence for ETS2 (NCBI Reference Sequence Database NM005239.5 ) preceded by a Kozak sequence was synthesized and cloned into a TOPO vector. This was linearized and a PCR amplicon generated, adding a T7 promoter and an AG initiation sequence (Phusion, NEB). A reverse complement (control) amplicon was also generated. These amplicons were used as templates for in vitro transcription using the HiScribe T7 mRNA Kit with CleanCap Reagent AG kit (NEB) according to the manufacturer’s instructions, but with substitution of N1-methyl-pseudouridine for uridine and methylcytidine for cytidine (both Stratech) to minimize non-specific cellular activation by the transfected mRNA. mRNA was purified using the MEGAclear Kit (Thermo Fisher Scientific) and polyadenylated using an Escherichia coli poly(A) polymerase (NEB) before further clean-up (MEGAclear), quantification and analysis of the product size (NorthernMax-Gly gel, Thermo Fisher Scientific). For optimizing overexpression conditions, GFP mRNA was produced using the same method. All primer sequences are provided in Supplementary Table 3 .
mRNA overexpression
Lipofectamine MessengerMAX (Thermo Fisher Scientific) was diluted in Opti-MEM (1:75 v/v), vortexed and incubated at room temperature for 10 min. IVT mRNA was then diluted in a fixed volume of Opti-MEM (112.5 µl per transfection), mixed with an equal volume of diluted Lipofectamine MessengerMAX and incubated for a further 5 min at room temperature. The transfection mix was then added dropwise to 2.5 × 10 6 M0 macrophages (precultured for 6 days in a six-well plate in antibiotic-free RPMI1640 macrophage medium containing M-CSF (50 ng ml −1 , Peprotech), with medium change on day 3). For GFP overexpression, cells were detached using Accutase 18 h after transfection and GFP expression was measured using flow cytometry. For ETS2 /control overexpression, either 250 ng or 500 ng mRNA was transfected and low-dose LPS (0.5 ng ml −1 ) was added 18 h after transfection, and cells were detached using Accutase 6 h later. Representative ETS2 expression in untransfected macrophages was obtained from previous data ( GSE193336 ). Differential H3K27ac ChIP–seq analysis in ETS2 -overexpressing macrophages was performed using 500 ng RNA transfection (see the ‘Code availability’ section).
Plink1.9 ( https://www.cog-genomics.org/plink/1.9/ ) was used to calculate a polygenic risk score (PRS) for patients in the IBD BioResource using 22 ETS2-regulated IBD-associated SNPs ( β coefficients from a previous study 3 ). Linear regression was used to compare PRSs with age at diagnosis, and logistic regression to estimate the effect of PRSs on IBD subphenotypes, including anti-TNF primary non-response (PNR), CD behaviour (B1 versus B2/B3), perianal disease and surgery. For variables with more than two levels (for example, CD location or UC location), ANOVA was used to investigate the relationship with PRS. For analyses of age at diagnosis, anti-TNF response and surgery, IBD diagnosis was included as a covariate.
Pathway analysis of 241 IBD-associated GWAS hits 3 was performed using SNPsea v.1.0.4 (ref. 34 ). In brief, linkage intervals were defined for every lead SNP based on the furthest correlated SNPs ( r 2 > 0.5 in 1000 Genomes, European population) and were extended to the nearest recombination hotspots with recombination rate > 3 cM per Mb. If no genes were present in this region, the linkage interval was extended up- and downstream by 500 kb (as long-range regulatory interactions usually occur within 1 Mb). Genes within linkage intervals were tested for enrichment within 7,660 pathways, comprising 7,658 GO Biological Pathways and two lists of ETS2-regulated genes (either those significantly downregulated after ETS2 disruption with gRNA1 or those significantly upregulated after ETS2 overexpression, based on a consensus list obtained from differential expression analysis including all samples and using donor and mRNA quantity as covariates). The analysis was performed using a single score mode: assuming that only one gene per linkage interval is associated with the pathway. A null distribution of scores for each pathway was performed by sampling identically sized random SNP sets matched on the number of linked genes (5,000,000 iterations). A permutation P value was calculated by comparing the score of the IBD-associated gene list with the null scores. An enrichment statistic was calculated using a standardized effect size for the IBD-associated score compared to the mean and s.e.m. of the null scores. Gene sets relating to the following IBD-associated pathways were extracted for comparison: NOD2 signalling (GO:0032495), integrin signalling (GO:0033627, GO:0033622), TNF signalling (GO:0033209, GO:0034612), intestinal epithelium (GO:0060729, GO:0030277), Th17 cells (GO:0072539, GO:0072538, GO:2000318), T cell activation (GO:0046631, GO:0002827), IL-10 signalling (GO:0032613, GO:0032733) and autophagy (GO:0061919, GO:0010506, GO:0010508, GO:1905037, GO:0010507). SNPs associated with PSC 5 , 87 , ankylosing spondylitis 4 , 87 , Takayasu arteritis 6 , 88 , 89 and schizophrenia 90 (as a negative control) were collated from the indicated studies and tested for enrichment in ETS2-regulated gene lists.
ETS2 co-expression
Genes co-expressed with ETS2 across 67 human monocyte/macrophage activation conditions (normalized data from GSE47189 ) were identified using the rcorr function in the Hmisc package in R.
13 C-glucose GC–MS
ETS2 -edited or unedited TPP macrophages were generated in triplicate for each donor and on day 6, the medium was removed, cells were washed with PBS, and new medium with labelled glucose was added. Labelled medium was as follows: RPMI1640 medium, no glucose (Thermo Fisher Scientific), 10% FBS (Thermo Fisher Scientific), GlutaMax (Thermo Fisher Scientific), 13 C-labelled glucose (Cambridge Isotype Laboratories). After 24 h, a timepoint selected from a time-course to establish steady-state conditions, the supernatants were snap-frozen and macrophages were detached by scraping. Macrophages were washed three times with ice-cold PBS, counted, resuspended in 600 µl ice-cold chloroform:methanol (2:1, v/v) and sonicated in a waterbath (3 times for 8 min). All of the extraction steps were performed at 4 °C as previously described 91 . The samples were analysed on the Agilent 7890B-7000C GC–MS system. Spitless injection (injection temperature of 270 °C) onto a DB-5MS (Agilent) was used, using helium as the carrier gas, in electron ionization mode. The initial oven temperature was 70 °C (2 min), followed by temperature gradients to 295 °C at 12.5 °C per min and to 320 °C at 25 °C per min (held for 3 min). The scan range was m / z 50–550. Data analysis was performed using in-house software MANIC (v.3.0), based on the software package GAVIN 92 . Label incorporation was calculated by subtracting the natural abundance of stable isotopes from the observed amounts. Total metabolite abundance was normalized to the internal standard (scyllo-inositol 91 ).
Roxadustat in TPP macrophages
ETS2- edited or unedited TPP macrophages were generated as described previously. On day 5 of culture, cells were detached (Accutase) and replated at a density of 10 5 cells per well in 96-well round-bottom plates in TPP medium containing roxadustat (FG-4592, 30 μM). After 12 h, cells were collected for functional assays and RNA-seq as described.
CUT&RUN
Precultured TPP macrophages were collected and processed immediately using the CUT&RUN Assay kit (Cell Signaling) according to the manufacturer’s instructions but omitting the use of ConA-coated beads. In brief, 5 × 10 5 cells per reaction were pelleted, washed and resuspended in antibody binding buffer. Cells were incubated with antibodies: anti-ETS2 (1:100, Thermo Fisher Scientific) or IgG control (1:20, Cell Signaling) for 2 h at 4 °C. After washing in digitonin buffer, cells were incubated with pA/G-MNase for 1 h at 4 °C. Cells were washed twice in digitonin buffer, resuspended in the same buffer and cooled for 5 min on ice. Calcium chloride was added to activate pA/G-MNase digestion (30 min, 4 °C) before the reaction was stopped and cells incubated at 37 °C for 10 min to release cleaved chromatin fragments. DNA was extracted from the supernatants using spin columns (Cell Signaling). Library preparation was performed using the NEBNext Ultra II DNA Library Prep Kit according to a protocol available at protocols.io ( https://doi.org/10.17504/protocols.io.bagaibse ). Size selection was performed using AMPure XP beads (Beckman Coulter) and the fragment size was assessed using the Agilent 2100 Bioanalyzer (High Sensitivity DNA kit). Indexed libraries were sequenced on the NovaSeq 6000 system (100 bp paired-end reads). Raw data were analysed using guidelines from the Henikoff laboratory 93 . In brief, paired-end reads were trimmed using Trim Galore and aligned to the human genome (GRCh37/hg19) using Bowtie2. BAM files were sorted, merged (technical and, where indicated, biological replicates), resorted and indexed using SAMtools. Picard was used to mark unmapped reads and SAMtools to remove these, re-sort and re-index. Bigwig files were created using the deepTools bamCoverage function. Processed data were initially analysed using the nf-core CUT&RUN pipeline v.3.0, using CPM normalization and default MACS2 parameters for peak calling. This analysis yielded acceptable quality metrics (including an average FRiP score of 0.23) but there was a high number of peaks with low fold enrichment (<4) over the control. More stringent parameters were therefore applied for peak calling (--qvalue 0.05 -f BAMPE --keep-dup all -B --nomodel) and we applied an irreproducible discovery rate (IDR; cut-off 0.001) to identify consistent peaks between replicates, implemented in the idr package in R (see the ‘Code availability’ section). Enrichment of binding motifs for ETS2 and other transcription factors expressed in TPP macrophages (cpm > 0.5) within consensus IDR peaks was calculated using TFmotifView 94 using global genomic controls. The overlap between consensus IDR peaks and the core promoter (−250bp to +35 bp from the transcription start site) and/or putative cis -regulatory elements of ETS2-regulated genes was assessed using differentially expressed gene lists after ETS2 disruption (gRNA1) or ETS2 overexpression (based on a consensus across mRNA doses, as described earlier). Putative cis -regulatory elements were defined as shared interactions (CHiCAGO score > 5) in monocyte and M0 and M1 macrophage samples from publicly available promoter-capture Hi-C data 61 . Predicted ETS2- and PU.1-binding sites were identified at the rs2836882 locus (chr21:40466150–40467450) using CisBP 95 (database 2.0, PWMs log odds motif model, default settings).
Intestinal scRNA-seq
Raw count data from colonic immune cells 41 (including healthy controls and Crohn’s disease) were downloaded from the Single Cell Portal ( https://singlecell.broadinstitute.org/single_cell ). Myeloid cell data were extracted for further analysis using the cell annotation provided. Raw data were preprocessed, normalized and variance-stabilized using Seurat (v.4) 96 . PCA and UMAP clustering was performed and clusters annotated using established markers and/or previous literature. Marker genes were identified using the FindAllMarkers function. Modular expression of ETS2-regulated genes (downregulated after ETS2 editing, gRNA1) was measured using the AddModuleScore function.
Spatial transcriptomics
Formalin-fixed paraffin-embedded sections (thickness, 5 μm) were cut from two PSC liver explants and two controls (healthy liver adjacent to tumour metastases), baked overnight at 60 °C and prepared for CosMx according to manufacturer’s instructions using 15 min target retrieval and 30 min protease digestion. Tissue samples were obtained through Tissue Access for Patient Benefit (TAP-B, part of the UCL-RFH Biobank) under research ethics approval: 16/WA/0289 (Wales Research Ethics Committee 4). One case and one control were included on each slide. The Human Universal Cell Characterization core panel (960 genes) was used, supplemented with 8 additional genes to improve identification of cells of interest: CD1D , EREG , ETS2 , FCN1 , G0S2 , LYVE1 , MAP2K1 , MT1G . Segmentation was performed using the CosMx Human Universal Cell Segmentation Kit (RNA), Human IO PanCK/CD45 Kit (RNA) and Human CD68 Marker, Ch5 (RNA). Fields of view (FOVs) were tiled across all available regions (221 control, 378 PSC) and cyclic fluorescence in situ hybridization was performed using the CosMx SMI (Nanostring) system. Data were preprocessed on the AtoMx Spatial Informatics Platform, with images segmented to obtain cell boundaries, transcripts assigned to single cells, and a transcript by cell count matrix was obtained 97 . Expression matrices, transcript coordinates, polygon coordinates, FOV coordinates and cell metadata were exported, and quality control, normalization and cell-typing were performed using InSituType 98 —an R package developed to extract all the information available in every cell’s expression profile. A semi-supervised strategy was used to phenotype cells, incorporating the Liver Human Cell Atlas reference matrix. Spatial analysis of macrophage phenotypes was performed according to proximity from cholangiocytes (anchor cell type). Radius and nearest-neighbour analyses were performed using PhenoptR ( https://akoyabio.github.io/phenoptr/ ) with macrophage distribution from cholangiocytes binned in 100 µm increments up to 500 µm. Nearest-neighbour analysis was performed to determine the distance from cholangiocytes to the nearest inflammatory and non-inflammatory macrophage and vice versa.
To generate overlay images, raw transcript and image (morphology 2D) data were exported from AtoMx. Overlays of selected ETS2-target genes ( CXCL8 , S100A9 , CCL2 , CCL5 ) and fluorescent morphology markers were generated using napari (v.0.4.17, https://napari.org/stable/index.html ) on representative FOVs: FOV287 (PSC with involved duct), FOV294 (PSC background liver) and FOV55 (healthy liver).
Chr21q22 disease datasets
Publicly available raw RNA-seq data from the affected tissues of chr21q22-associated diseases (and controls from the same experiment) were downloaded from the GEO: IBD macrophages ( GSE123141 ), PSC liver ( GSE159676 ), ankylosing spondylitis synovium ( GSE41038 ). Reads were trimmed, filtered and aligned as described earlier. For each disease dataset, a ranked list of genes was obtained by differential expression analysis between cases and controls using limma with voom transformation. For IBD macrophages, only IBD samples with active disease were included. fGSEA using ETS2-regulated gene lists was performed as described.
LINCS signatures
A total of 31,027 lists of downregulated genes after exposure of a cell line to a small molecule was obtained from the NIH LINCS database 7 (downloaded in January 2021). These were used as gene sets for fGSEA (as described) with a ranked list of genes obtained by differential expression analysis between ETS2 -edited and unedited TPP macrophages (gRNA1) using limma with voom transformation and donor as a covariate. Drug classes for gene sets with FDR-adjusted P < 0.05 were manually assigned on the basis of known mechanisms of action.
MEK inhibition in TPP macrophages
TPP macrophages were generated as described previously. On day 4 of culture, PD-0325901 (0.5 μM, Sigma-Aldrich) or vehicle (DMSO) was added. Cells were collected on day 6 and RNA was extracted and sequenced as described.
Colonic biopsy explant culture
During colonoscopy, intestinal mucosal biopsies (6 per donor) were collected from ten patients with IBD (seven patients with ulcerative colitis, three patients with Crohn’s disease). All had endoscopically active disease and were not receiving immunosuppressive or biologic therapies. All biopsies were collected from a single inflamed site. All patients provided written informed consent. Ethical approval was provided by the London–Brent Regional Ethics Committee (21/LO/0682). Biopsies were collected into Opti-MEM and, within 1 h, were weighed and placed in pairs onto a Transwell insert (Thermo Fisher Scientific), designed to create an air–liquid interface 99 , in a 24-well plate. Each well contained 1 ml medium and was supplemented with either DMSO (vehicle control), PD-0325901 (0.5 μM) or infliximab (10 μg ml −1 ; MSD). Medium was as follows: Opti-MEM I (Gibco), GlutaMax (Thermo Fisher Scientific), 10% FBS (Thermo Fisher Scientific), MEM non-essential amino acids (Thermo Fisher Scientific), 1% sodium pyruvate (Thermo Fisher Scientific), 1% penicillin–streptomycin (Thermo Fisher Scientific) and 50 μg ml −1 gentamicin (Merck). After 18 h, the supernatants and biopsies were snap-frozen. The supernatant cytokine concentrations were quantified using the LEGENDplex Human Inflammation Panel (BioLegend). RNA was extracted from biopsies and libraries were prepared as described earlier ( n = 9, RNA from one donor was too degraded). Sequencing was performed on the NovaSeq 6000 system (100 bp paired-end reads). Data were processed as described earlier and GSVA was performed for ETS2-regulated genes and biopsy-derived signatures of IBD-associated inflammation 46 .
Chr21q22 genotypes in archaic humans
Using publicly available genomes from seven Neanderthal individuals 100 , 101 , 102 , 103 , one Denisovan individual 104 , and one Neanderthal and Denisovan F1 individual 105 , genotypes were called at the disease-associated chr21q22 candidate SNPs from the respective BAM files using bcftools mpileup with base and mapping quality options -q 20 -Q 20 -C 50 and using bcftools call -m -C alleles, specifying the two alleles expected at each site in a targets file (-T option). From the resulting .vcf file, the number of reads supporting the reference and alternative alleles was extracted and stored in the ‘DP4’ field.
Inference of Relate genealogy at rs2836882
Genome-wide genealogies, previously inferred for samples of the Simons Genome Diversity Project 106 using Relate 107 , 108 ( https://reichdata.hms.harvard.edu/pub/datasets/sgdp/ ), were downloaded from https://www.dropbox.com/sh/2gjyxe3kqzh932o/AAAQcipCHnySgEB873t9EQjNa?dl=0 . Using the inferred genealogies, the genealogy at rs2836882 (chr21:40466570) was plotted using the TreeView module of Relate.
Data presentation
The following R packages were used to create figures: GenomicRanges 109 , EnhancedVolcano 110 , ggplot2 (ref. 111 ), gplots 112 , karyoploteR 113 .
Statistical methodology
Statistical methods used in MPRA analysis, fGSEA and SNPsea are described above. For other analyses, comparison of continuous variables between two groups was performed using Wilcoxon matched-pairs tests (paired) or Mann–Whitney U -tests (unpaired) for nonparametric data or a t -tests for parametric data. Comparison against a hypothetical value was performed using Wilcoxon signed-rank tests for nonparametric data or one-sample t -tests for parametric data. A Shapiro–Wilk test was used to confirm normality. Two-sided tests were used as standard unless a specific hypothesis was being tested. Sample sizes are provided in the main text and figure captions.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The datasets produced in this study are accessible at the following repositories: MPRA (GEO: GSE229472 ), RNA-seq data of ETS2 or chr21q22-edited TPP macrophages (EGA: EGAD00001011338 ), RNA-seq data of ETS2 overexpression (EGA: EGAD00001011341 ), RNA-seq data of MEK-inhibitor-treated TPP macrophages (EGA: EGAD00001011337 ), H3K27ac ChIP–seq data in TPP macrophages (EGA: EGAD00001011351 ), ATAC–seq and H3K27ac ChIP–seq data in ETS2 -overexpressing or -edited macrophages (EGA: EGAD50000000154 ), ETS2 CUT&RUN data (EGA: EGAD00001011349 ), biopsy RNA-seq data (EGA: EGAD00001011333 ). MetaboLights: Metabolomics (MTBLS7665). The counts table for CosMx is provided at Zenodo ( https://zenodo.org/records/10707942 ) 114 . The phenotype and genotype data used for the PRS analysis are available on application to the IBD Bioresource ( https://www.ibdbioresource.nihr.ac.uk/ ). Source data are provided with this paper.
Code availability
Code to reproduce analyses are available at GitHub ( https://github.com/JamesLeeLab/chr21q22_manuscript ; https://github.com/chr1swallace/ibd-ets2-analysis ; https://github.com/qzhang314/PRS_IBD_subpheno ) 114 . Final code is deposited at Zenodo ( https://zenodo.org/records/10707942 ).
Miller, F. W. The increasing prevalence of autoimmunity and autoimmune diseases: an urgent call to action for improved understanding, diagnosis, treatment, and prevention. Curr. Opin. Immunol. 80 , 102266 (2023).
Article CAS PubMed Google Scholar
Dowden, H. & Munro, J. Trends in clinical success rates and therapeutic focus. Nat. Rev. Drug Discov. 18 , 495–496 (2019).
de Lange, K. M. et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat. Genet. 49 , 256–261 (2017).
Article PubMed PubMed Central Google Scholar
International Genetics of Ankylosing Spondylitis Consortium et al. Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat. Genet. 45 , 730–738 (2013).
Article Google Scholar
Ji, S. G. et al. Genome-wide association study of primary sclerosing cholangitis identifies new risk loci and quantifies the genetic relationship with inflammatory bowel disease. Nat. Genet. 49 , 269–273 (2017).
Ortiz-Fernandez, L. et al. Identification of susceptibility loci for Takayasu arteritis through a large multi-ancestral genome-wide association study. Am. J. Hum. Genet. 108 , 84–99 (2021).
Stathias, V. et al. LINCS Data Portal 2.0: next generation access point for perturbation-response signatures. Nucleic Acids Res. 48 , D431–D439 (2020).
Harrison, R. K. Phase II and phase III failures: 2013–2015. Nat. Rev. Drug Discov. 15 , 817–818 (2016).
Claussnitzer, M. et al. A brief history of human disease genetics. Nature 577 , 179–189 (2020).
Article CAS PubMed PubMed Central ADS Google Scholar
King, E. A., Davis, J. W. & Degner, J. F. Are drug targets with genetic support twice as likely to be approved? Revised estimates of the impact of genetic support for drug mechanisms on the probability of drug approval. PLoS Genet. 15 , e1008489 (2019).
Cader, M. Z. et al. C13orf31 (FAMIN) is a central regulator of immunometabolic function. Nat. Immunol. 17 , 1046–1056 (2016).
Article CAS PubMed PubMed Central Google Scholar
Murthy, A. et al. A Crohn’s disease variant in Atg16l1 enhances its degradation by caspase 3. Nature 506 , 456–462 (2014).
Article CAS PubMed ADS Google Scholar
Hnisz, D. et al. Super-enhancers in the control of cell identity and disease. Cell 155 , 934–947 (2013).
Park, M. D., Silvin, A., Ginhoux, F. & Merad, M. Macrophages in health and disease. Cell 185 , 4259–4279 (2022).
Kugathasan, S. et al. Loci on 20q13 and 21q22 are associated with pediatric-onset inflammatory bowel disease. Nat. Genet. 40 , 1211–1215 (2008).
Xue, J. et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity 40 , 274–288 (2014).
Kuo, D. et al. HBEGF + macrophages in rheumatoid arthritis induce fibroblast invasiveness. Sci. Transl. Med. 11 , eaau8587 (2019).
Melnikov, A. et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat. Biotechnol. 30 , 271–277 (2012).
Nerlov, C. & Graf, T. PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev. 12 , 2403–2412 (1998).
Minderjahn, J. et al. Mechanisms governing the pioneering and redistribution capabilities of the non-classical pioneer PU.1. Nat. Commun. 11 , 402 (2020).
Martinez, L. A. Mutant p53 and ETS2, a tale of reciprocity. Front. Oncol. 6 , 35 (2016).
Wei, G. et al. Activated Ets2 is required for persistent inflammatory responses in the motheaten viable model. J. Immunol. 173 , 1374–1379 (2004).
Zhao, J., Huang, K., Peng, H. Z. & Feng, J. F. Protein C-ets-2 epigenetically suppresses TLRs-induced interleukin 6 production in macrophages. Biochem. Biophys. Res. Commun. 522 , 960–964 (2020).
Chung, S. W., Chen, Y. H. & Perrella, M. A. Role of Ets-2 in the regulation of heme oxygenase-1 by endotoxin. J. Biol. Chem. 280 , 4578–4584 (2005).
Quinn, S. R. et al. The role of Ets2 transcription factor in the induction of microRNA-155 (miR-155) by lipopolysaccharide and its targeting by interleukin-10. J. Biol. Chem. 289 , 4316–4325 (2014).
Ma, X. et al. Ets2 suppresses inflammatory cytokines through MAPK/NF-κB signaling and directly binds to the IL-6 promoter in macrophages. Aging 11 , 10610–10625 (2019).
Aperlo, C., Pognonec, P., Stanley, E. R. & Boulukos, K. E. Constitutive c-ets2 expression in M1D + myeloblast leukemic cells induces their differentiation to macrophages. Mol. Cell. Biol. 16 , 6851–6858 (1996).
Henkel, G. W. et al. PU.1 but not ets-2 is essential for macrophage development from embryonic stem cells. Blood 88 , 2917–2926 (1996).
Mittal, M., Siddiqui, M. R., Tran, K., Reddy, S. P. & Malik, A. B. Reactive oxygen species in inflammation and tissue injury. Antioxid. Redox Signal. 20 , 1126–1167 (2014).
Kelly, B. & O’Neill, L. A. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. 25 , 771–784 (2015).
Martin, J. C. et al. Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell 178 , 1493–1508 (2019).
Peloquin, J. M. et al. Characterization of candidate genes in inflammatory bowel disease-associated risk loci. JCI Insight 1 , e87899 (2016).
Sazonovs, A. et al. Large-scale sequencing identifies multiple genes and rare variants associated with Crohn’s disease susceptibility. Nat. Genet. 54 , 1275–1283 (2022).
Slowikowski, K., Hu, X. & Raychaudhuri, S. SNPsea: an algorithm to identify cell types, tissues and pathways affected by risk loci. Bioinformatics 30 , 2496–2497 (2014).
Cramer, T. et al. HIF-1α is essential for myeloid cell-mediated inflammation. Cell 112 , 645–657 (2003).
Tannahill, G. M. et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 496 , 238–242 (2013).
Shiratori, R. et al. Glycolytic suppression dramatically changes the intracellular metabolic profile of multiple cancer cell lines in a mitochondrial metabolism-dependent manner. Sci. Rep. 9 , 18699 (2019).
Landt, S. G. et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 22 , 1813–1831 (2012).
Basuyaux, J. P., Ferreira, E., Stehelin, D. & Buttice, G. The Ets transcription factors interact with each other and with the c-Fos/c-Jun complex via distinct protein domains in a DNA-dependent and -independent manner. J. Biol. Chem. 272 , 26188–26195 (1997).
Sevilla, L. et al. Bcl-XL expression correlates with primary macrophage differentiation, activation of functional competence, and survival and results from synergistic transcriptional activation by Ets2 and PU.1. J. Biol. Chem. 276 , 17800–17807 (2001).
Kong, L. et al. The landscape of immune dysregulation in Crohn’s disease revealed through single-cell transcriptomic profiling in the ileum and colon. Immunity 56 , 444–458 (2023).
Chapuy, L. et al. Two distinct colonic CD14 + subsets characterized by single-cell RNA profiling in Crohn’s disease. Mucosal Immunol. 12 , 703–719 (2019).
Newman, J. A., Cooper, C. D., Aitkenhead, H. & Gileadi, O. Structural insights into the autoregulation and cooperativity of the human transcription factor Ets-2. J. Biol. Chem. 290 , 8539–8549 (2015).
Liu, H. et al. ERK differentially regulates Th17- and T reg -cell development and contributes to the pathogenesis of colitis. Eur. J. Immunol. 43 , 1716–1726 (2013).
Koboziev, I., Karlsson, F., Zhang, S. & Grisham, M. B. Pharmacological intervention studies using mouse models of the inflammatory bowel diseases: translating preclinical data into new drug therapies. Inflamm. Bowel Dis. 17 , 1229–1245 (2011).
Article PubMed Google Scholar
Argmann, C. et al. Biopsy and blood-based molecular biomarker of inflammation in IBD. Gut 72 , 1271–1287 (2023).
Malle, L. et al. Autoimmunity in Down’s syndrome via cytokines, CD4 T cells and CD11c + B cells. Nature 615 , 305–314 (2023).
Feagan, B. G. et al. Guselkumab plus golimumab combination therapy versus guselkumab or golimumab monotherapy in patients with ulcerative colitis (VEGA): a randomised, double-blind, controlled, phase 2, proof-of-concept trial. Lancet Gastroenterol. Hepatol. 8 , 307–320 (2023).
Friedrich, M. et al. IL-1-driven stromal-neutrophil interactions define a subset of patients with inflammatory bowel disease that does not respond to therapies. Nat. Med. 27 , 1970–1981 (2021).
Klesse, L. J. et al. The use of MEK inhibitors in neurofibromatosis type 1-associated tumors and management of toxicities. Oncologist 25 , e1109–e1116 (2020).
Zou, Y., Carbonetto, P., Wang, G. & Stephens, M. Fine-mapping from summary data with the “sum of single effects” model. PLoS Genet. 18 , e1010299 (2022).
1000 Genomes Consortium. A map of human genome variation from population-scale sequencing. Nature 467 , 1061–1073 (2010).
Kerimov, N. et al. A compendium of uniformly processed human gene expression and splicing quantitative trait loci. Nat. Genet. 53 , 1290–1299 (2021).
Fairfax, B. P. et al. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science 343 , 1246949 (2014).
Quach, H. et al. Genetic adaptation and Neandertal admixture shaped the immune system of human populations. Cell 167 , 643–656 (2016).
Chen, L. et al. Genetic drivers of epigenetic and transcriptional variation in human immune cells. Cell 167 , 1398–1414 (2016).
Nedelec, Y. et al. Genetic ancestry and natural selection drive population differences in immune responses to pathogens. Cell 167 , 657–669 (2016).
Alasoo, K. et al. Shared genetic effects on chromatin and gene expression indicate a role for enhancer priming in immune response. Nat. Genet. 50 , 424–431 (2018).
Wallace, C. A more accurate method for colocalisation analysis allowing for multiple causal variants. PLoS Genet. 17 , e1009440 (2021).
Bourges, C. et al. Resolving mechanisms of immune-mediated disease in primary CD4 T cells. EMBO Mol. Med. 12 , e12112 (2020).
Javierre, B. M. et al. Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell 167 , 1369–1384 (2016).
Hanzelmann, S., Castelo, R. & Guinney, J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 14 , 7 (2013).
Peters, J. E. et al. Insight into genotype-phenotype associations through eQTL mapping in multiple cell types in health and immune-mediated disease. PLoS Genet. 12 , e1005908 (2016).
Conant, D. et al. Inference of CRISPR edits from Sanger trace data. CRISPR J. 5 , 123–130 (2022).
Kalita, C. A. et al. QuASAR-MPRA: accurate allele-specific analysis for massively parallel reporter assays. Bioinformatics 34 , 787–794 (2018).
de Santiago, I. et al. BaalChIP: Bayesian analysis of allele-specific transcription factor binding in cancer genomes. Genome Biol. 18 , 39 (2017).
Corces, M. R. et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14 , 959–962 (2017).
Calderon, D. et al. Landscape of stimulation-responsive chromatin across diverse human immune cells. Nat. Genet. 51 , 1494–1505 (2019).
Reske, J. J., Wilson, M. R. & Chandler, R. L. ATAC-seq normalization method can significantly affect differential accessibility analysis and interpretation. Epigenet. Chromatin 13 , 22 (2020).
Article CAS Google Scholar
Brown, A. C. et al. Comprehensive epigenomic profiling reveals the extent of disease-specific chromatin states and informs target discovery in ankylosing spondylitis. Cell Genom. 3 , 100306 (2023).
Lienhard, M., Grimm, C., Morkel, M., Herwig, R. & Chavez, L. MEDIPS: genome-wide differential coverage analysis of sequencing data derived from DNA enrichment experiments. Bioinformatics 30 , 284–286 (2014).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 , 139–140 (2010).
Randzavola, L. O. et al. EROS is a selective chaperone regulating the phagocyte NADPH oxidase and purinergic signalling. eLife 11 , e76387 (2022).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37 , 907–915 (2019).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25 , 2078–2079 (2009).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30 , 923–930 (2014).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43 , e47 (2015).
Korotkevich, G. et al. Fast gene set enrichment analysis. Preprint at bioRxiv https://doi.org/10.1101/060012 (2021).
Qing, G. et al. Single-cell RNA sequencing revealed CD14 + monocytes increased in patients with Takayasu’s arteritis requiring surgical management. Front. Cell Dev. Biol. 9 , 761300 (2021).
Smillie, C. S. et al. Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell 178 , 714–730 (2019).
Gao, K. M. et al. Human nasal wash RNA-Seq reveals distinct cell-specific innate immune responses in influenza versus SARS-CoV-2. JCI Insight 6 , e152288 (2021).
Yang, Q. et al. The interaction of macrophages and CD8 T cells in bronchoalveolar lavage fluid is associated with latent tuberculosis infection. Emerg. Microbes Infect. 12 , 2239940 (2023).
Reyes, M. et al. An immune-cell signature of bacterial sepsis. Nat. Med. 26 , 333–340 (2020).
Mulder, K. et al. Cross-tissue single-cell landscape of human monocytes and macrophages in health and disease. Immunity 54 , 1883–1900 (2021).
Cassetta, L. et al. Human tumor-associated macrophage and monocyte transcriptional landscapes reveal cancer-specific reprogramming, biomarkers, and therapeutic targets. Cancer Cell 35 , 588–602 (2019).
Zernecke, A. et al. Integrated single-cell analysis-based classification of vascular mononuclear phagocytes in mouse and human atherosclerosis. Cardiovasc. Res. 119 , 1676–1689 (2023).
Ellinghaus, D. et al. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat. Genet. 48 , 510–518 (2016).
Renauer, P. A. et al. Identification of susceptibility loci in IL6, RPS9/LILRB3, and an intergenic locus on chromosome 21q22 in Takayasu arteritis in a genome-wide association study. Arthritis Rheumatol. 67 , 1361–1368 (2015).
Terao, C. et al. Genetic determinants and an epistasis of LILRA3 and HLA-B*52 in Takayasu arteritis. Proc. Natl Acad. Sci. USA 115 , 13045–13050 (2018).
Trubetskoy, V. et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature 604 , 502–508 (2022).
Bussi, C. et al. Lysosomal damage drives mitochondrial proteome remodelling and reprograms macrophage immunometabolism. Nat. Commun. 13 , 7338 (2022).
Behrends, V., Tredwell, G. D. & Bundy, J. G. A software complement to AMDIS for processing GC-MS metabolomic data. Anal. Biochem. 415 , 206–208 (2011).
Meers, M. P., Bryson, T. D., Henikoff, J. G. & Henikoff, S. Improved CUT&RUN chromatin profiling tools. eLife 8 , e46314 (2019).
Leporcq, C. et al. TFmotifView: a webserver for the visualization of transcription factor motifs in genomic regions. Nucleic Acids Res. 48 , W208–W217 (2020).
Weirauch, M. T. et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158 , 1431–1443 (2014).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184 , 3573–3587 (2021).
He, S. et al. High-plex imaging of RNA and proteins at subcellular resolution in fixed tissue by spatial molecular imaging. Nat. Biotechnol. 40 , 1794–1806 (2022).
Danaher, P. et al. InSituType: likelihood-based cell typing for single cell spatial transcriptomics. Preprint at bioRxiv https://doi.org/10.1101/2022.10.19.512902 (2022).
Vadstrup, K. et al. Validation and optimization of an ex vivo assay of intestinal mucosal biopsies in Crohn’s disease: reflects inflammation and drug effects. PLoS ONE 11 , e0155335 (2016).
Prufer, K. et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505 , 43–49 (2014).
Article PubMed ADS Google Scholar
Prufer, K. et al. A high-coverage Neandertal genome from Vindija Cave in Croatia. Science 358 , 655–658 (2017).
Article PubMed PubMed Central ADS Google Scholar
Hajdinjak, M. et al. Reconstructing the genetic history of late Neanderthals. Nature 555 , 652–656 (2018).
Mafessoni, F. et al. A high-coverage Neandertal genome from Chagyrskaya Cave. Proc. Natl Acad. Sci. USA 117 , 15132–15136 (2020).
Meyer, M. et al. A high-coverage genome sequence from an archaic Denisovan individual. Science 338 , 222–226 (2012).
Slon, V. et al. The genome of the offspring of a Neanderthal mother and a Denisovan father. Nature 561 , 113–116 (2018).
Mallick, S. et al. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature 538 , 201–206 (2016).
Speidel, L., Forest, M., Shi, S. & Myers, S. R. A method for genome-wide genealogy estimation for thousands of samples. Nat. Genet. 51 , 1321–1329 (2019).
Speidel, L. et al. Inferring population histories for ancient genomes using genome-wide genealogies. Mol. Biol. Evol. 38 , 3497–3511 (2021).
Lawrence, M. et al. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 9 , e1003118 (2013).
Blighe, K. et al. EnhancedVolcano: publication-ready volcano plots with enhanced colouring and labeling. Bioconductor https://doi.org/10.18129/B9.bioc.EnhancedVolcano (2023).
Hadley, W. Ggplot2 (Springer, 2016).
Warnes, G. et al. gplots: various R programming tools for plotting data. CRAN https://CRAN.R-project.org/package=gplots (2022).
Gel, B. & Serra, E. karyoploteR: an R/Bioconductor package to plot customizable genomes displaying arbitrary data. Bioinformatics 33 , 3088–3090 (2017).
Stankey, C. T. et al. Data for ‘A disease-associated gene desert directs macrophage inflammation through ETS2’. Zenodo https://doi.org/10.5281/zenodo.10707942 (2024).
Download references
Acknowledgements
We thank the members of the Lee laboratory, K. Slowikowski and A. Kaser for discussions; G. Stockinger, C. Vinuesa, C. Swanton, R. Patani and C. Reis e Sousa for reading the manuscript; C. Cheshire and the staff at the Francis Crick Institute Advanced Sequencing Facility and Flow Cytometry STP for technical support; L. Lucaciu for help with patient recruitment; RFH PITU nurses for assistance obtaining infliximab; the members of Tissue Access for Patient Benefit (TAP-B) for providing liver samples; NIHR BioResource volunteers for their participation; and the NIHR BioResource centres, NHS Blood and Transplant, and NHS staff for their contributions. This work was supported by Crohn’s and Colitis UK (M2018-3), the Wellcome Trust (Sir Henry Wellcome Fellowship to L.S., 220457/Z/20/Z; Investigator Award to P.S., 217223/Z/19/Z; Senior Fellowship to C.W., WT220788; Clinical Research Career Development Fellowship to M.Z.C., 222056/Z/20/Z; Wellcome-Beit Prize Clinical Career Development Fellowship to D.C.T., 206617/A/17/A; and Intermediate Clinical Fellowship to J.C.L., 105920/Z/14/Z), and the Francis Crick Institute, which receives its core funding from Cancer Research UK (CC2219, FC001595), the UK Medical Research Council (CC2219, FC001595) and the Wellcome Trust (CC2219, FC001595). L.M.H. is supported by the Charité–Universitätsmedizin Berlin and the Berlin Institute of Health Charité (Clinician-Scientist Program); A.J.C. by the Medical Research Council (MR/V029711/1); A.L. by a Lord Kelvin/Adam Smith Leadership Grant; A.H.S. by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIH, R01:AR070148); N.B.J. by Cancer Research UK (C55370/A25813); T.Z. by the Chinese Scholarship Council (202308060128); A.Q. by the NIHR UCLH/UCL BRC; J.C.K. by Versus Arthritis (program grant, 20773), Janssen Oxford Translational fellowships and NIHR Oxford BRC; P.S. by the European Molecular Biology Organisation, the Vallee Foundation and the European Research Council (852558); C.W. by the Medical Research Council (MC UU 00002/4), GSK, MSD and the NIHR Cambridge BRC (BRC-1215-20014); and D.C.T. by the Sidharth Burman endowment. J.C.L. is a Lister Institute Prize Fellow. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. Experimental schematics in Figs. 1d , 2a and 3a and Extended Data Figs. 3a , 4a,b,e and 7g,h were created using BioRender. For the purpose of open access, the authors have applied a CC BY public copyright licence to any author accepted manuscript version arising from this submission.
Open Access funding provided by The Francis Crick Institute.
Author information
These authors contributed equally: C. T. Stankey, C. Bourges, L. M. Haag
Authors and Affiliations
Genetic Mechanisms of Disease Laboratory, The Francis Crick Institute, London, UK
C. T. Stankey, C. Bourges, T. Turner-Stokes, A. P. Piedade, I. Papa, E. C. Parkes & J. C. Lee
Department of Immunology and Inflammation, Imperial College London, London, UK
C. T. Stankey, T. Turner-Stokes & L. O. Randzavola
Washington University School of Medicine, St Louis, MO, USA
C. T. Stankey
Division of Gastroenterology, Infectious Diseases and Rheumatology, Charité–Universitätsmedizin Berlin, Berlin, Germany
Department of Gastroenterology, Royal Free Hospital, London, UK
C. Palmer-Jones, A. P. Rochford, C. D. Murray & J. C. Lee
Institute for Liver and Digestive Health, Division of Medicine, University College London, London, UK
C. Palmer-Jones, F. Saffioti, D. Thorburn, A. P. Rochford, C. D. Murray & J. C. Lee
Metabolomics STP, The Francis Crick Institute, London, UK
M. Silva dos Santos & J. I. MacRae
Genomics of Inflammation and Immunity Group, Human Genetics Programme, Wellcome Sanger Institute, Hinxton, UK
Wolfson Wohl Cancer Centre, School of Cancer Sciences, University of Glasgow, Glasgow, UK
A. J. Cameron, A. Legrini, T. Zhang, C. S. Wood & N. B. Jamieson
NanoString Technologies, Seattle, WA, USA
F. N. New & P. Divakar
Ancient Genomics Laboratory, The Francis Crick Institute, London, UK
L. Speidel & P. Skoglund
Genetics Institute, University College London, London, UK
Wellcome Centre for Human Genetics, University of Oxford, Oxford, UK
A. C. Brown & J. C. Knight
The Sheila Sherlock Liver Centre, Royal Free Hospital, London, UK
A. Hall, F. Saffioti & D. Thorburn
Department of Cellular Pathology, Royal Free Hospital, London, UK
A. Hall & A. Quaglia
Cambridge Institute of Therapeutic Immunology and Infectious Disease, University of Cambridge, Cambridge, UK
W. Edwards, M. Z. Cader, C. Wallace & D. C. Thomas
Department of Internal Medicine, Division of Rheumatology, Marmara University, Istanbul, Turkey
H. Direskeneli
Systemic Autoimmunity Branch, NIAMS, National Institutes of Health, Bethesda, MD, USA
P. C. Grayson
Department of Rheumatology, Zhongshan Hospital, Fudan University, Shanghai, China
Division of Rheumatology, Department of Medicine, University of Pennsylvania, Philadelphia, PA, USA
P. A. Merkel
Division of Epidemiology, Department of Biostatistics, Epidemiology and Informatics, University of Pennsylvania, Philadelphia, PA, USA
Department of Physiology, Istanbul University, Istanbul Faculty of Medicine, Istanbul, Turkey
G. Saruhan-Direskeneli
Division of Rheumatology, Department of Pediatrics, University of Pittsburgh, Pittsburgh, PA, USA
A. H. Sawalha
Division of Rheumatology and Clinical Immunology, Department of Medicine, University of Pittsburgh, Pittsburgh, PA, USA
Lupus Center of Excellence, University of Pittsburgh, Pittsburgh, PA, USA
Department of Immunology, University of Pittsburgh, Pittsburgh, PA, USA
Department of Biomedical and Clinical Sciences, Milan University, Milan, Italy
E. Tombetti
Internal Medicine and Rheumatology, ASST FBF-Sacco, Milan, Italy
UCL Cancer Institute, London, UK
Chinese Academy of Medical Sciences Institute, Nuffield Department of Medicine, University of Oxford, Oxford, UK
J. C. Knight
NIHR Comprehensive Biomedical Research Centre, Oxford, UK
Experimental Histopathology STP, The Francis Crick Institute, London, UK
M. Green & E. Nye
Department of Medicine, University of Cambridge, Cambridge, UK
M. Z. Cader & D. C. Thomas
MRC Biostatistics Unit, Cambridge Institute of Public Health, Cambridge, UK
You can also search for this author in PubMed Google Scholar
Contributions
Conceptualization: J.I.M., N.B.J., P.S., M.Z.C., C.W., D.C.T. and J.C.L. Methodology: C.T.S., C.B., M.S.d.S., M.G., E.N., J.I.M., C.W. and J.C.L. Software: C.B., M.S.d.S., Q.Z., A.J.C., A.L., T.Z., C.S.W., L.S., J.I.M., N.B.J., P.S., C.W. and J.C.L. Investigation: C.T.S., C.B., L.M.H., T.T.-S., A.P.P., I.P., M.S.d.S., L.O.R., A.C.B., E.C.P., W.E., M.G., C.D.M. and J.C.L. Resources: C.T.S., C.B., C.P.-J., A.H., F.S., A.Q., D.T., A.P.R., C.D.M. and J.C.L. Formal analysis: C.T.S., C.B., M.S.d.S., Q.Z., A.J.C., A.L., T.Z., F.N.N., L.S., P.D., C.W. and J.C.L. Writing—original draft: C.T.S., C.B. and J.C.L. Writing—review and editing: all of the authors. Funding acquisition: J.C.L. Supervision: J.C.K., J.I.M., N.B.J., P.S., C.W., D.C.T. and J.C.L.
Corresponding author
Correspondence to J. C. Lee .
Ethics declarations
Competing interests.
C.T.S., C.B. and J.C.L. are listed as co-inventors on a patent application related to this work. C.W. holds a part-time position at GSK. GSK had no role in the design or conduct of this study. F.N.N. and P.D. are employees and shareholders of NanoString Technologies. NanoString had no role in the design or conduct of this study. The other authors declare no competing interests.
Peer review
Peer review information.
Nature thanks Joachim Schultze and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended data fig. 1 colocalisation between genetic associations at chr21q22..
a . Example comparison of genetic associations at chr21q22: IBD and ETS2 eQTL in unstimulated monocytes. Plot adapted from locuscomparer. b . Tukey box-and-whisker plot depicting ETS2 expression stratified by rs2836882 genotype in unstimulated monocytes (AA, n = 39; AG, n = 142; GG, n = 233) 54 . P -value is as reported in index study. c . Radar plot of representative colocalization results for the indicated genetic associations compared to IBD. Posterior probability of independent causal variants, PP.H3, dark blue; posterior probability of shared causal variant, PP.H4, light blue. PP.H4 > 0.5 was used to call colocalisation (denoted by dashed line). Labels are coloured according to class of data (indicated in the key). Asterisks denote colocalisation. Data sources are: IBD 3 , PSC 5 , AS 4 , Takayasu Arteritis 6 , BLUEPRINT 56 , Fairfax 54 , Quach 55 , Nedelec 57 , Alasoo 58 .
Extended Data Fig. 2 CRISPR-Cas9 editing of the chr21q22 locus and ETS2 in monocytes.
a . Cas9 gRNAs were designed to flank the chr21q22 enhancer region at the indicated sites. b . Representative bioanalyzer trace of PCR-amplified target region following monocyte CRISPR/Cas9 editing with an equimolar mix of RNPs containing 5′ and 3′ chr21q22 gRNAs. Example editing efficiency calculation shown. c . Editing efficiency at the chr21q22 locus. Mean enhancer deletion: 42.4% (n = 11). d . Location and sequence of gRNAs used to disrupt ETS2 . e . ETS2 editing efficiency. gRNA1 (mean), 89.7% (n = 31); gRNA2 (mean), 78.6% (n = 14). f . ETS2 expression (relative to NTC) following CRISPR/Cas9 editing, measured by qPCR (housekeeping gene PPIA ; equivalent results with other housekeeping genes; n = 10). g . Viability following monocyte nucleofection with Cas9 RNPs and macrophage differentiation. Mean values: NTC, 97.9%; gRNA1: 98.3%; gRNA2, 98.6% (n = 6). h . Expression of myeloid lineage markers following ETS2 editing and TPP differentiation (n = 5). Gating strategy shown in Supplementary Information Fig. 2 . i . GSVA enrichment scores for 67 different monocyte/macrophage activation conditions to identify stimuli that phenocopy CD14+ monocytes/macrophages from IBD patients. j . Chromatin accessibility in ETS2-edited versus unedited inflammatory macrophages (n = 3). k . Enhancer activity (H3K27ac) in ETS2-edited versus unedited inflammatory macrophages (n = 3). P values calculated using edgeR (two-sided) in j , k . Red points denote adjusted P -value (P adj ) < 0.1, grey points NS. Error bars are mean±SEM in c , e - h . * P < 0.05. NTC: non-targeting control.
Extended Data Fig. 3 Optimization of MPRA and mRNA overexpression in primary human macrophages.
a . Schematic of MPRA. A library of oligonucleotides (each containing a genomic sequence and unique barcode, separated by restriction enzyme sites) is cloned into a pGL4.10 M cloning vector. A promoter and reporter gene are inserted using directional cloning. The resulting plasmids are transfected into primary human macrophages (TPP) and RNA is extracted after 24 h. Barcode abundance in cellular mRNA and input DNA library are quantified by high-throughput sequencing, and mRNA barcode counts are normalized to corresponding counts in DNA library to assess expression-modulating activity. b . Identification of suitable promoters for MPRA in TPP macrophages. TPP macrophages were transfected with reporter vectors, each with GFP expression under the control of a different promoter. GFP expression was quantified by flow cytometry after 24 h. c . Adapted MPRA vector for use in primary human macrophages, containing RSV promoter. d . Heatmap showing pairwise correlation of expression-modulating activity of all constructs between donors. e . Principal component analysis of element counts (sum of barcodes tagging same genomic sequence) in mRNA from TPP macrophages (n = 8 donors; red) and four replicates of DNA vector (black). f . Primary human macrophages (M0) were transfected with different quantities of GFP mRNA using Lipofectamine MessengerMAX. GFP expression was quantified by flow cytometry 18 h after transfection. g . Cytokine secretion following ETS2 overexpression. Plot shows relative cytokine concentrations in macrophage supernatants ( ETS2 relative to control) following transfection with 500 ng mRNA (n = 11). Error bars are mean±SEM. One-sample t -test (two-tailed) * P < 0.05, ** P < 0.01. The diagram in a was created using BioRender.
Extended Data Fig. 4 Molecular effects of allelic variation at rs2836882 .
a . Schematic of PU.1 ChIP-genotyping assay to assess allele-specific PU.1 binding at rs2836882 in human macrophages. b . Schematic of standard curve generation by TaqMan genotyping various pre-defined ratios of risk and non-risk containing DNA sequences. c . Standard curve generated using different allelic ratios of 200-nt DNA geneblocks centred on either the major (risk) or minor (non-risk) rs2836882 allele. d . Allele-specific PU.1 binding at rs2836882 in TPP macrophages (one-sample t -test, two-sided, n = 5). Error bars represent mean±95%CI. e . Schematic of PU.1 MPRA-ChIP assay to assess allele-specific PU.1 binding at individual SNPs within chr21q22 enhancer. f . Allele-specific PU.1 binding at SNPs within chr21q22 enhancer in TPP macrophages. Data represents the allelic ratio of normalized PU.1 binding for constructs centred on the SNP allele from the MPRA library (fixed-effects meta-analysis of QuASAR-MPRA results, two-sided, n = 6). Box represents median (IQR), whiskers represent minima and maxima. g . Allele-specific ATAC-seq reads at rs2836882 in two deeply sequenced heterozygous TPP macrophage datasets (left: 154.7 million non-duplicate paired-end reads, right: 165.4 million non-duplicate paired-end reads). h . H3K27ac ChIP-seq data from risk (red) or non-risk (blue) allele homozygotes at rs2836882 (n = 4). i . Rank Ordering of Super-Enhancers (ROSE) analysis of H3K27ac ChIP-seq data from TPP macrophages from major (left) and minor (right) allele homozygotes. Dashed line denotes inflection point of curve, with enhancers above this point being denoted as super-enhancers. Red points indicate rs2836882 -containing chr21q22 enhancer. SE, super-enhancer. The diagrams in a , b and e were created using BioRender.
Extended Data Fig. 5 Functional effects of the chr21q22 enhancer.
a . Extracellular ROS production by unedited (NTC), chr21q22-edited, and ETS2 g1-edited TPP macrophages, quantified by chemiluminescence. Points represent relative area under curve for edited versus unedited cells (Wilcoxon signed-rank test, two-sided; n = 6). b . Cytokine secretion from inflammatory macrophages following deletion of the chr21q22 enhancer. Heatmap shows relative cytokine concentrations in the supernatants of chr21q22-edited TPP macrophages versus unedited (NTC) cells (Wilcoxon signed rank test, one-sided; n = 7). c . Representative flow cytometry histograms demonstrating phagocytosis of fluorescently-labelled zymosan particles by chr21q22-edited and unedited (NTC) TPP macrophages. d . Phagocytosis index for unedited and chr21q22-edited TPP macrophages, calculated as proportion of positive cells multiplied by mean fluorescence intensity of positive cells. Plot shows relative phagocytosis index for chr21q22-edited cells versus unedited cells (Wilcoxon signed-rank test two-sided; n = 7). e . Enrichment of differentially-expressed genes following deletion of the disease-associated chr21q22 locus (upregulated genes, top; downregulated genes, bottom) in ETS2 -edited versus unedited macrophages. P adj , FDR-adjusted P -value (two-sided). f . Tukey box-and-whisker plot depicting quantitative PCR of selected ETS2-target genes in resting (M0) macrophages from minor and major allele homozygote IBD patients (n = 22, expression normalized to PPIA and scaled to minimum 0, maximum 1). Mann-Whitney test (one-sided). * P < 0.05, ** P < 0.01, *** P < 0.001.
Extended Data Fig. 6 Polygenic Risk Score of 22 ETS2-regulated IBD-associated genes.
a . Summary of IBD BioResource cohorts used for PRS analysis. b . Association between PRS and age at diagnosis. c . Association between PRS and extent of ulcerative colitis (E1, proctitis; E2, left-sided; E3, extensive colitis). d . Association between PRS and Crohn’s disease location (L1, ileal; L2, colonic; L3, ileocolonic). L2 is associated with a milder disease phenotype. e . Association between PRS and perianal involvement in Crohn’s disease. f . Association between PRS and Crohn’s disease behaviour (B1, inflammatory; B2, stricturing; B3, fistulating). B2 and B3 represent more aggressive, complicated forms of Crohn’s disease. g . Association between PRS and response to anti-TNFα in Crohn’s disease and ulcerative colitis (PR, primary responder; PNR, primary non-responder). h . Association between PRS and need for surgery in Crohn’s disease and ulcerative colitis. Overall, higher PRS was associated with: earlier age at diagnosis, ileal or ileocolonic forms of Crohn’s disease, B2/B3 Crohn’s disease behaviour, and increased need for surgery in IBD. Analysis in b performed using linear regression. Analyses in c - h performed using logistic regression (with diagnosis as covariate in g and h ). SNPs included in PRS are listed in Extended Data Table 1 . i . Plot of enrichment statistic (standardized effect size) against statistical significance from SNPsea analysis of genes tagged by 241 IBD SNPs within ETS2 -regulated genes (red) and known IBD pathways (black). j . SNPsea analyses of SNPs associated with PSC, ankylosing spondylitis, Takayasu’s arteritis or Schizophrenia (negative control) within lists of ETS2-regulated genes–either upregulated by ETS2 overexpression, downregulated by ETS2 disruption, or downregulated following chr21q22 deletion (all FDR < 0.05). Dashed line denotes P < 0.05.
Extended Data Fig. 7 Effects of modulating ETS2.
a and b . Changes in total metabolite abundance ( a ) and percentage of label incorporation from 13 C-glucose ( b ) following ETS2 editing in TPP macrophages (n = 6). Colour depicts median log2 fold-change in ETS2 -edited macrophages relative to unedited macrophages (transfected with non-targeting control RNPs; NTC). Bold black border indicates P < 0.05 (Wilcoxon signed rank test, two-sided). c . Heatmap summarizing metabolic changes following ETS2 disruption. Colour depicts median log2 fold-change in ETS2 g1-edited cells relative to unedited cells (Wilcoxon signed rank test, two-sided, * P < 0.05). d . Phagocytosis index in unedited (NTC) and ETS2 -edited TPP macrophages treated with roxadustat (ROX) or vehicle. Phagocytosis index is calculated as proportion of positive cells multiplied by mean fluorescence intensity of positive cells (488 nm channel). Data normalized to phagocytosis index in unedited cells (n = 5). e . Extracellular ROS production by unedited (NTC) and ETS2 -edited TPP macrophages treated with ROX or vehicle – quantified using a chemiluminescence assay. Data represent log2 fold-change of area under curve (AUC) normalized to unedited (NTC) TPP macrophages (n = 5). f . TFmotifView enrichment results for motifs of transcription factors expressed in TPP macrophages (CPM > 0.5) within ETS2 CUT&RUN peaks. Results shown for all significantly enriched transcription factors (Bonferroni P value < 0.05, two-sided) with motifs in more than 10% peaks. g . Schematic of experiment to assess how ETS2 disruption affects the activity of the chr21q22 ETS2 enhancer in inflammatory (TPP) macrophages. h . Schematic of experiment to assess how ETS2 overexpression affects the activity of the chr21q22 ETS2 enhancer in resting (M0) macrophages. i . Normalized H3K27ac ChIP-seq read counts (edgeR fitted values) from chr21:40,465,000-40,470,000 in experiments depicted in g (left) and h (right) (edgeR P values, two-sided, n = 3 for each). Error bars in d and e represent mean±SEM. The diagrams in g and h were created using BioRender.
Extended Data Fig. 8 The transcriptional signature of ETS2 is detectable in affected tissues from chr21q22-linked diseases.
a . ETS2 expression in scRNA-seq clusters of myeloid cells from Crohn’s disease and healthy controls (upper panel). Relative contributions of single cells from Crohn’s disease or healthy controls to individual clusters (same UMAP dimensions as for combined analysis). b . Overlay of CosMx morphology 2D image data and raw transcripts of selected ETS2 target genes. Fluorescent morphology markers alone (top row), CXCL8 (cyan) and S1009A (yellow) transcripts (middle row), CCL5 (cyan) and CCL2 (yellow) transcripts (bottom row). Columns are representative examples of PSC with diseased ducts (left), PSC with uninflamed background liver (centre), and healthy liver (right). Size marker (white) on every field of view (FOV) denotes 50 µm. c . Gene set enrichment analysis (fGSEA) of genes downregulated following chr21q22 enhancer deletion or ETS2 disruption (gRNA1 or gRNA2) within intestinal macrophages from patients with active IBD (compared to control intestinal macrophages, n = 20; left), ankylosing spondylitis synovium (compared to control synovium, n = 15; centre), and PSC liver biopsies (compared to control liver biopsies, n = 17; right). P adj , FDR-adjusted P -value (two-sided).
Extended Data Fig. 9 Effect of MEK1/2 inhibition on ETS2- regulated genes.
a - c . Gene set enrichment analysis (fGSEA) in MEK1/2 inhibitor-treated TPP macrophages showing enrichment of gene sets upregulated (upper panel) or downregulated (lower panel) following ETS2 or chr21q22 editing (MEK1/2 inhibited using PD-0325901, 0.5 µM). Gene sets obtained from differential gene expression analysis (limma using voom transformation) following ETS2 disruption with gRNA1 ( a ), gRNA2 ( b ), or following chr21q22 deletion ( c ). d . fGSEA in intestinal biopsies from IBD patients showing enrichment of gene sets downregulated following ETS2 or chr21q22 editing in MEK inhibitor-treated biopsies. Upregulated gene sets were not enriched. e . Proportion and pathway analysis of MEK inhibitor-induced differentially expressed genes that have no evidence for being ETS2 targets in macrophages (incorporating differential expression from knockout or overexpression experiments and promoter / regulatory element binding from ETS2 CUT&RUN). P adj , FDR-adjusted P -value (two-sided).
Extended Data Fig. 10 Geographic distribution and history of rs2836882 .
a . rs2836882 allele frequency in modern global populations (data from 1000 Genomes Project, plotted using Geography of Genetic Variants browser: https://popgen.uchicago.edu/ggv/ ). b . Genotypes of candidate SNPs at chr21q22 (99% credible set) in archaic humans (Neanderthals and Denisovans). Colour depicts the proportion of reads containing ALT alleles, with a value close to 0 consistent with a homozygous REF (risk) genotype, a value close to 1 consistent with a homozygous ALT (non-risk) genotype, and an intermediate value indicating a potential heterozygous genotype. Number in each cell indicates the number of reads at that SNP in the indicated sample. Putative causal variant highlighted in red. c . Inferred genealogy of the age of the rs2836882 polymorphism – analysed using Relate. The diagram in a was created using the Geography of Genetic Variants browser.
Supplementary information
Supplementary figures.
Supplementary Fig. 1: uncropped Western blots from Fig. 2d. Two lanes were run for each sample: one lane to blot for vinculin and the NADPH oxidase components gp91phox, gp65 and p22phox, and one lane to blot for vinculin and the chaperone protein EROS. After transfer, the membranes were cut to blot for individual targets. Supplementary Fig. 2: example gating strategy. Example gating strategy for MPRA and macrophage phenotyping. Macrophages were gated by FSC-A/SSC-A and singlets were gated by FSC-A/FSC-H. Live cells were gated (and viability was quantified) using Live/Dead Fixable Aqua Dead Cell Stain.
Reporting Summary
Supplementary tables.
Supplementary Table 1: differentially expressed genes in primary macrophages after ETS2 or chr21q22 CRISPR–Cas9 editing. Supplementary Table 2: differentially expressed genes in primary macrophages after ETS2 overexpression. Supplementary Table 3: the primers and gRNA sequences used in this study.
Peer Review File
Source data, source data fig. 1, source data fig. 2, source data fig. 3, source data fig. 4, source data fig. 5, source data extended data fig. 1, source data extended data fig. 2, source data extended data fig. 3, source data extended data fig. 4, source data extended data fig. 5, source data extended data fig. 6, source data extended data fig. 7, source data extended data fig. 8, source data extended data fig. 9, source data extended data fig. 10, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Stankey, C.T., Bourges, C., Haag, L.M. et al. A disease-associated gene desert directs macrophage inflammation through ETS2. Nature 630 , 447–456 (2024). https://doi.org/10.1038/s41586-024-07501-1
Download citation
Received : 17 April 2023
Accepted : 01 May 2024
Published : 05 June 2024
Issue Date : 13 June 2024
DOI : https://doi.org/10.1038/s41586-024-07501-1
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
By submitting a comment you agree to abide by our Terms and Community Guidelines . If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing: Translational Research newsletter — top stories in biotechnology, drug discovery and pharma.

PTFSpot: Deep co-learning on transcription factors and their binding regions attains impeccable universality in plants
- Find this author on Google Scholar
- Find this author on PubMed
- Search for this author on this site
- For correspondence: [email protected]
- Info/History
- Supplementary material
- Preview PDF
Unlike animals, variability in transcription factors (TF) and their binding regions (TFBR) across the plants species is a major problem which most of the existing TFBR finding software fail to tackle, rendering them hardly of any use. This limitation has resulted into underdevelopment of plant regulatory research and rampant use of Arabidopsis like model species, generating misleading results. Here we report a revolutionary transformers based deep-learning approach, PTFSpot, which learns from TF structures and their binding regions co-variability to bring a universal TF-DNA interaction model to detect TFBR with complete freedom from TF and species specific models' limitations. During a series of extensive benchmarking studies over multiple experimentally validated data, it not only outperformed the existing software by >30% lead, but also delivered consistently >90% accuracy even for those species and TF families which were never encountered during model building process. PTFSpot makes it possible now to accurately annotate TFBRs across any plant genome even in the total lack of any TF information, completely free from the bottlenecks of species and TF specific models.
Competing Interest Statement
The authors have declared no competing interest.
Material and Methods, Result and Discussion Sections are updated; Some Figures revised; Supplemental files updated.
View the discussion thread.
Supplementary Material
Thank you for your interest in spreading the word about bioRxiv.
NOTE: Your email address is requested solely to identify you as the sender of this article.

Citation Manager Formats
- EndNote (tagged)
- EndNote 8 (xml)
- RefWorks Tagged
- Ref Manager
- Tweet Widget
- Facebook Like
- Google Plus One
Subject Area
- Bioinformatics
- Animal Behavior and Cognition (5387)
- Biochemistry (12153)
- Bioengineering (9099)
- Bioinformatics (30038)
- Biophysics (15418)
- Cancer Biology (12541)
- Cell Biology (17986)
- Clinical Trials (138)
- Developmental Biology (9717)
- Ecology (14559)
- Epidemiology (2067)
- Evolutionary Biology (18720)
- Genetics (12521)
- Genomics (17166)
- Immunology (12265)
- Microbiology (28919)
- Molecular Biology (11988)
- Neuroscience (62958)
- Paleontology (462)
- Pathology (1929)
- Pharmacology and Toxicology (3354)
- Physiology (5165)
- Plant Biology (10763)
- Scientific Communication and Education (1704)
- Synthetic Biology (2989)
- Systems Biology (7516)
- Zoology (1688)
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Springer Nature - PMC COVID-19 Collection

Transcription and Qualitative Methods: Implications for Third Sector Research
Caitlin mcmullin.
Department of Politics & Society, Aalborg University, Aalborg, Denmark
While there is a vast literature that considers the collection and analysis of qualitative data, there has been limited attention to audio transcription as part of this process. In this paper, I address this gap by discussing the main considerations, challenges and implications of audio transcription for qualitative research on the third sector. I present a framework for conducting audio transcription for researchers and transcribers, as well as recommendations for writing up transcription in qualitative research articles.
Introduction
The field of third sector studies is inherently interdisciplinary, with studies from political science, management, sociology and social work, among others. Within the field of research, a large percentage (between 40–80%) of studies employ qualitative methods such as interviews, focus groups and ethnographic observations (von Schnurbein et al., 2018 ). In order to ensure rigor, qualitative researchers devote considerable time to developing interview guides, consent forms and coding frameworks. While there is a vast literature that considers the collection and the analysis of qualitative data, there has been comparatively limited attention paid to audio transcription, which is the conversion of recorded audio material into a written form that can be analyzed. Despite advances made in qualitative methodologies and increasing attention to positionality, subjectivity and reliability in qualitative data analysis, the transcription of interviews and focus groups is often presented uncritically as a direct conversion of recorded audio to text. As technology to facilitate transcription improves, many researchers have shifted to using voice-to-text software and companies that employ AI rather than human transcription. These technological advances in transcription, along with shifts in the way that research is undertaken (for example, increasingly via video conferencing as a result of the COVID-19 pandemic), mean that the need to critically reflect upon the place of transcription in third sector research is more urgent.
In this article, I explore the place of transcription in qualitative research, with a focus on the importance of this process for third sector researchers. The article is structured as follows. First, I review the qualitative methods literature on audio transcription and the key themes that arise. Next, I report on a review undertaken of recent qualitative research articles in Voluntas and the way that authors discuss transcription in these articles. Finally, I propose a framework for qualitative third sector researchers to include transcription as part of their research design and elements to consider in including descriptions of the transcription process in writing up qualitative research.
Audio Transcription: What We Know
At a basic level, transcription refers to the transformation of recorded audio (usually spoken word) into a written form that can be used to analyze a particular phenomenon or event (Duranti, 2006 ). For many qualitative researchers, transcription has become a fairly taken-for-granted aspect of the research process. In this section, I review the methods literature on the process of audio (and video) transcription as part of qualitative research on the third sector, focusing on three key areas—how transcription is undertaken, epistemological and ethical considerations, and the role of technology.
Qualitative research and transcription
While quantitative research seeks to explain, generalize and predict patterns through the analysis of variables, qualitative research questions are more interested in understanding and interpreting the socially constructed world around us (Bryman, 2016 ). This means that data are collected through documents, observation and interviews, and the latter are often recorded in order to analyze these as documents. For third sector research, recordings are most commonly made of interviews and focus groups, but may also be of meetings, events and other activities to ensure that researchers do not have to rely on their power of recall or scribbled notes.
Transcription is a notoriously time-consuming and often tedious task which can take between three hours and over eight hours to transcribe one hour of audio, depending on typing speed. Transcription is not, however, a mechanical process where the written document becomes an objective record of the event—indeed, written text varies from the spoken word in terms of syntax, word choice and accepted grammar (Davidson, 2009 ). The transcriber therefore has to make subjective decisions throughout about what to include (or not), whether to correct mistakes and edit grammar and repetitions. This has been described as a spectrum between “naturalized” transcription (or “intelligent verbatim”) which adapts the oral to written norms, and “denaturalized” transcription (“full verbatim”), where everything is left in, including utterances, mistakes, repetitions and all grammatical errors (Bucholtz, 2000 ).
While some contend that denaturalized transcription is more ‘accurate’, the same can equally be argued for naturalized, as it allows the transcriber to omit occasions when, for instance, an individual mis-speaks and corrects themselves, thereby allowing the transcriber to record closer to what was intended and how the interviewee might have portrayed themselves in a written form. As Lapadat ( 2000 , p. 206) explains, “Spoken language is structured and accomplished differently than written text, so when talk is re-presented as written text, it is not surprising that readers draw on their knowledge of written language to evaluate it.” Other nonverbal cues, such as laughter, tone of voice (e.g. sarcasm, frustration, emphasis) and the use or omission of punctuation, can also drastically alter the meaning or intention of what an individual says. In addition, the transcriber must make decisions about how much contextual information to include, such as interruptions, crosstalk and inaudible segments (Lapadat, 2000 ). Because of the range of types of research that employ qualitative methods, there is no single set of rules for transcription but rather these decisions must be based on the research questions and approach.
Epistemological and Ethical Considerations
Because the researcher (or external transcriber) must make these decisions as they translate audio into written text, transcription is an inherently interpretative and political act, influenced by the transcriber’s own assumptions and biases (Jaffe, 2007 ). Every choice that the transcriber makes therefore shapes how the research participant is portrayed and determines what knowledge or information is relevant and valuable and what is not. Indeed, two transcribers may hear differently and select relevant spoken material differently (Stelma & Cameron, 2007 ). As Davidson ( 2009 ) notes (and as I explore in further detail in the next section), despite being a highly interpretive process, transcription is frequently depicted using positivist norms of knowledge creation.
Transcription also involves potential ethical considerations and dilemmas. When working with disadvantaged communities, deciding how to depict research participants in written text can highlight the challenges of ethical representation. As Kvale ( 1996 , pp. 172–3) notes, “Be mindful that the publication of incoherent and repetitive verbatim interview transcripts may involve an unethical stigmatization of specific persons or groups of people”. Oliver et al. ( 2005 ) similarly demonstrate how transcribers must make decisions about how to represent participants’ use of slang, colloquialisms and accents in ways that are accurate but also respectful of the respondent’s intended meaning. Some researchers decide to send finished transcriptions to interviewees for approval in order to honor commitments to fully informed consent, to ensure transcription accuracy or in some cases as a means to address the balance of power between the researcher and interviewee. As Mero-Jaffe ( 2011 ) describes, on the one hand, this may empower interviewees to control the way that they are portrayed in the research. On the other hand, Mero-Jaffe found that seeking transcript approval from interviewees sometimes increased their embarrassment at the way that their statements appear in text. This may be especially problematic with full verbatim transcriptions.
Technology and Transcription
As technology improves and AI becomes increasingly able to create written text from recorded audio, researchers might ask—is human transcription even necessary? New options in Computer Assisted Qualitative Data Analysis Software (CAQDAS) such as NVivo, Atlas.ti and MAXQDA give qualitative researchers the option to forgo audio-to-text transcription altogether, and instead engage in live coding of audio or video files. Using this method, researchers first watch or listen to recordings to code for nonverbal cues, followed by a stage of note taking and coding based on pre-defined themes and matching these with time codes and nonverbal cues. Finally, researchers then transcribe specific quotes of interest from the recording (Parameswaran et al., 2020 ). This process may improve immersion in the data and allow researchers to account for dynamics that are often lost in complete audio-to-text transcription, such as group interactions and nonverbal communication.
There is a considerable need to develop the evidence base on the role of AI in transcription for qualitative research, with many important publications that consider the issue (e.g. Gibbs et al., 2002 ; Markle et al., 2011 ) out-of-date given the swift rate of change in AI technologies. Over the last few years, voice and speech recognition technologies have improved dramatically and may now be able to provide researchers with “good enough” first drafts of transcripts (Bokhove & Downey, 2018 ), providing certain conditions are in place (e.g. limited number of speakers and excellent audio quality). Using these technologies can save researchers time and money. As a result of the COVID-19 pandemic, many qualitative researchers are now undertaking interviews over Zoom or other video conferencing apps, which is a trend that may continue beyond the pandemic (Dodds & Hess, 2020 ). Zoom offers AI live transcription options, which benefits from the generally clear audio quality of a video conference, compared to in-person interviews where there is a greater chance of audio interference and background noise that may be undetected in the moment.
While AI may offer a cheaper and quicker alternative to human transcription, these transcripts will need to be meticulously checked by the researcher to ensure accuracy, fill in missing details or edit for context and readability. Using cloud-based AI transcription services also raises potential ethical concerns about data protection and confidentiality (Da Silva, 2021 ). There are numerous subjective decisions made in the course of creating a transcription that AI is unable to process, such as where to include punctuation, which words to include or exclude (such as filler words, hesitations, etc.) and how to denote things such as interruptions, hesitations and nonverbal cues. Voice-to-text software is also generally less accurate in discerning multiple voices or different accents (Bokhove & Downey, 2018 ). Several studies have considered how researchers/transcribers can use voice recognition software to listen and repeat the spoken text of an interview into software as a shortcut to traditional typing transcription (Matheson, 2007 ; Tilley, 2003 ), but the above shortcomings and cautions apply.
Transcription and Third Sector Research
Transcription matters for third sector research because qualitative research methodologies make up a large percentage of studies undertaken on nonprofits—as much as 40–80% of research published in this field (Igalla et al., 2019 ; Laurett & Ferreira, 2018 ; von Schnurbein et al., 2018 ). Audio transcription is particularly important for third sector research for several reasons. In conducting qualitative research (which aims to produce rich, rigorous description) and as third sector researchers (who study organizations that seek to improve society and who may be working with traditionally disenfranchised or disadvantaged communities), we have a particular ethical obligation to ensure that our research provides an accurate depiction of our participants’ lives and the organizations with which they are involved.
However, transcription is perhaps the most underacknowledged aspect of the qualitative research process, and this is also evident in the way that transcription is discussed in research articles. In order to survey the current depiction of the transcription process in third sector research, I undertook a review of the 212 most recent papers in Voluntas that include the word ‘interview’ to explore how qualitative research articles discuss transcription as part of their methodology. 1 Of these papers, 79 were deemed not applicable (because they were quantitative research papers that mentioned interviews in another context, or used the word interview to denote the administering of a structured questionnaire, or systematic review papers reporting on other research). This left 133 articles which were analyzed to explore the extent to which transcription was described—if at all—as part of the research methodology. 2
The analysis (illustrated in Fig. 1 ) found that 41% of papers employing interviews as a research method did not mention transcription at all, while 11% mentioned transcripts but not the process of transcription. It was not clear from these whether or not interviews were recorded or if researchers relied upon written notes taken during interviews, or how information from the oral interview was converted into analyzable text. The most common discussion of transcription (19%) was a simple sentence along the lines of “interviews were recorded and transcribed”, while 26% gave some further information including who undertook the transcription (the researcher(s), a research assistant or a commercial company) or that the interviews were transcribed ‘verbatim’ (with none explaining what they mean by this term). These findings are not dissimilar to a study of qualitative research in nursing, where it was found that 66% of articles reporting solely that interviews were transcribed, and the remaining articles indicated only “full” or “verbatim” to clarify the process (Wellard & McKenna, 2001 ). I also surveyed the first authors’ departmental affiliations/field of study to gauge any differences between academic fields (Table (Table1) 1 ) although there were not considerable differences.

Transcription in Voluntas qualitative articles
Description of transcription and field of first authors
| Field | Transcription not discussed or passive mention of transcripts | Discussion of transcription | Interviews not recorded | Total |
|---|---|---|---|---|
| Business, management & economics | 13 | 17 | 1 | 31 |
| 42% | 55% | 3% | ||
| Political science and international development | 18 | 8 | 0 | 26 |
| 69% | 31% | |||
| Sociology | 8 | 12 | 0 | 20 |
| 40% | 60% | |||
| Public policy & public administration | 12 | 6 | 0 | 18 |
| 67% | 33% | |||
| Social work | 4 | 7 | 1 | 12 |
| 33% | 58% | 8% | ||
| Other or field unclear | 14 | 10 | 2 | 26 |
| 54% | 38% | 8% | ||
The fact that over half of the Voluntas articles using interviews as a research method make no mention of the transcription process is a problem for transparency in qualitative research. This tendency may be a symptom of the fact that qualitative researchers face greater challenges in academic publishing that disadvantage longer from, in-depth qualitative research to fit within prescribed word limits (Moravcsik, 2014 ). In researchers’ efforts to ensure that qualitative research meets requirements for transparency, rigor and reliability, efforts are concentrated on descriptions of case and participant selection and data analysis while transcription as the conduit between data collection and analysis remains unproblematized. This emphasis reflects the growing influence of positivist views of validity. Ignoring the subjective decisions and theoretical perspectives that determine the creation of a transcript therefore inadvertently presupposes a positivist stance on the objective nature of data which is inconsistent with qualitative methodologies.
A Framework for Undertaking and Reporting on Transcription
As shown in the previous section, there is currently widespread neglect of transcription as part of interpretive qualitative research on the third sector. In this section, I present key elements for third sector researchers to consider in regard to transcription, both to ensure rigor as part of the qualitative research process and in writing up qualitative research, drawing upon examples of good practice from previous research in Voluntas. These recommendations are based on a review of the literature as well as my personal experience as a qualitative researcher, qualitative methods teacher, and professional transcriber.
Before Transcribing: Ethics and Data Management
All decisions regarding research design, data collection and data management should be made at the beginning of a qualitative research project when applying for ethical/IRB approval from one’s university, and this includes transcription. At this stage, the researcher should confirm with their university whether they have a budget for transcription. Undertaking ethical qualitative research means ensuring standards of transparency, informed consent, confidentiality and protection of the data obtained from the research (Blaxter et al., 2001 ). Increasing concerns about data protection and legislation such as GDPR in the European Union have prompted many universities to institute strict rules about where research data can be stored. Some universities do not allow the use of certain cloud servers, such as Dropbox. These considerations should be taken into account when deciding how to undertake and record interviews (Da Silva, 2021 )—for instance, if you are recording using your mobile phone, it is important to be sure you know whether recordings automatically upload to the cloud. For this reason, it may be preferable to use a traditional digital recorder so you can manually download the files to your computer and know exactly where everything is saved.
Before Transcribing: The Interview
Before transcription can even be considered, researchers must ensure that they have a suitable audio recording, which begins with the interview itself—whenever possible, interviews should be conducted in a quiet environment without background noise or interruptions and the audio recording device should be placed close enough to the respondent to pick up their voice clearly. While recording interviews with a mobile phone has become increasingly common and easy, using a backup recording device is always a good idea to mitigate against flat batteries, full memory cards, and human error. If recording with your mobile phone, it’s also critical to remember to place it on airplane mode/‘do not disturb’ for the duration of the interview.
To Transcribe or Not to Transcribe?
While transcription from audio recordings is considered standard practice in qualitative research (Tracy, 2019 ), it is not the only way of undertaking qualitative interviews, and it is important to note that there are many reasons why it may not be desirable, appropriate or possible to record interviews at all. In relation to third sector research, this is most commonly the case in community-based research, research with political elites or research in challenging environments. One article explained that they did not record interviews because: “In sectors marked by fear, intimidation, and strong security apparatuses, recording devices would almost certainly have led to self-censorship and limited our access.” (Atia & Herrold, 2018 , p. 1046). Similarly, researchers may be unable to record in community settings because of sub-optimal recording conditions (e.g. meeting outside, noisy environments, etc.) or because using recording device makes participants uncomfortable or reinforces power relations between the researcher and participants (Quintanilha et al., 2015 ).
If researchers decide not to comprehensively transcribe recordings, or decide not to record qualitative fieldwork at all, this should be noted and explained in relation to methods. Other methods of notetaking and analysis may be more suited to certain types of ethnographic research, such as reflexive journaling (Halcomb & Davidson, 2006 ), or Systematic and Reflexive Interviewing and Reporting—a process by which a researcher and research assistant jointly interview participants and write their own reports that include observations and analyses, which are collaboratively analyzed (Loubere, 2017 ).
How to Transcribe?
Traditionally, transcribers used foot pedals to play, rewind and fast forward tape recordings while they typed. Now that audio files are digital, several free and low cost programs are available (such as Express Scribe and oTranscribe) that let transcribers set up hot keys to perform the same actions without having to navigate away from their transcript document.
The degree of detail to include in transcripts should be decided upon before interviews are transcribed. This is important because previous research has demonstrated that the format selected for transcription significantly impacts how the researcher interprets the data (Mishler, 2003 ; Packer, 2017 ). There is no one best or “most accurate” style of transcription, but rather, a researcher should consider the particular theoretical background and research questions of the study in order to determine where on the scale of full verbatim to intelligent verbatim is most appropriate for the study. Because third sector research is most commonly associated with social science and business disciplines rather than linguistics, it will rarely be necessary or appropriate to employ the conventions of conversation analysis or extreme levels of denaturalized transcription (Bucholtz, 2000 ). Indeed, it might most frequently be appropriate to employ a version of naturalized/intelligent verbatim, so that any participants’ quotes included in written works are more ‘readable’ and do not include excessive repetitions or verbal fillers such as ‘um’.
If the researcher determines that naturalized or intelligent verbatim transcription is the most appropriate for their study, several considerations should be heeded in order to ensure that meaning is not distorted or lost. First, indications of laughter, nonverbal cues (such as sighs, huffs, finger-snaps, sobbing or even blowing raspberries) should be included if these convey important meaning. Other considerations of how to transcribe may be based more on personal preference and the ability to produce a document that is easily analyzable in the researcher’s chosen medium. For instance, wide margins on one side can be useful for researchers who choose to analyze their data on paper or in Microsoft Word, while other more flowing templates will work better to import into software such as NVivo. It can also be useful to include time stamps for unclear or inaudible statements, or at regular intervals (e.g. every minute) which makes it much easier to check a transcript against the original audio.
Who Transcribes?
As discussed in the consideration of qualitative studies, the prevalence of the passive voice when reporting on transcription (i.e. “interviews were transcribed”) obscures the important distinction of who undertook the transcription. If the researcher transcribes recordings themselves, then it is generally acceptable to assume the coherence between the research approach and approach to transcription, as well as the researcher’s confidence that the written transcript is an accurate record of the event/interview that took place. If, however, the researchers choose to outsource transcription to a research assistant or commercial transcription company, then care should be taken to give detailed and thorough instructions about the elements described above. The researcher should also spot check transcripts for accuracy, fill in any missed words/inaudibles and ensure that the transcription document fulfils their expectations in regard to level of verbatim, style and formatting.
Ideally, transcribers should be hired who have specialist knowledge of the subject matter and familiarity with the accents or dialect of the speakers. They should be provided with a key information about the project, such as the research questions, important terms and acronyms. Lapadat ( 2000 ) provides several useful suggestions when hiring transcribers in order to ensure transcription quality and increase rigor. First, rather than fully outsourcing transcription, the researchers can transcribe some interviews or portions of interviews themselves in order to provide an example for transcribers and develop a transcription protocol. Another option when employing research assistants to transcribe interviews is to include them directly in the interviews (either as a co-interviewer or observer), so they have direct involvement in the research and context.
Finally, when working with external transcribers it can also be valuable to encourage transcribers to keep memos of the transcription process or contextual observations and impressions that may not come through in the written text. For instance, does the interviewee sound tired, frustrated, distracted or nervous? Does the interviewer interrupt the respondent frequently (which the transcriber may choose to edit for readability)? Or did the interview take place somewhere public, like a cafe, which may have made the respondent more guarded? Such information is often lost, particularly in projects that involve multiple research team members (for instance, a PI, multiple interviewers, research assistants and/or professional transcribers).
Writing about Transcription
Due to limited space or word limits, it is not typically possible or desirable to include all of the above details in research articles. Instead, at a minimum, researchers should include who transcribed the audio recordings as part of a commitment to ethical and transparent qualitative research. If this was done by anyone other than the researchers, authors should ideally describe the measures taken to ensure accuracy (developing a protocol for transcribers, spot checking, proofreading, sending transcripts to interviewees if appropriate) and ethical considerations (such as data protection and confidentiality).
Second, researchers should indicate the type of transcription—whether selective (pulling out relevant quotes and themes, or transcribing just the ‘gist’), intelligent verbatim/naturalized or full verbatim/denaturalized. The choice of type of transcription should align to the researcher’s epistemological position and theoretical framework.
Finally, researchers should include any other subjective decision-making that took place during the transcription process, in much the same way that researchers are encouraged to be transparent about their subjectivity and positionality in undertaking interviews and analysis of qualitative data (McCorkel & Myers, 2003 ). This may include information about selecting the level of verbatim, working with external transcribers, feedback from interviewees on transcripts or efforts to ensure accuracy of transcripts and coherence with the research approach.
The following quotes provide good examples of how to write about transcription:
The interviews, which were conducted in the native language of the interviewees by six female Hebrew-Arabic-speaking interviewers, were recorded, translated, and transcribed verbatim. […] Immediately following the interview, each interviewer transcribed and translated her interviews into Hebrew. In this manner, we sought to achieve a translation that was as close as possible to the interviewer’s insights regarding the participants, and we regarded the interviewers as active agents in the creation of knowledge. (Yanay-Ventura et al., 2020 , p. 6) Three Spanish speaking investigators transcribed all of the interviews from audio recording devices, checked each other’s transcription for accuracy, and analyzed the interviews using thematic analysis (Braun & Clarke, 2006 ). The transcribers observed the focus groups and took notes on participants’ voices and other identifying traits to help the transcription process go more smoothly. Researchers aided the transcribers in this regard by asking participants at the beginning of the focus groups to introduce themselves using a pseudonym and briefly remark upon how they preferred to spend their time. (Schwingel et al., 2017 , p. 170)
In both of these examples, the authors treat the process of transcription as part of the broader research process, rather than as an automatic conversion of audio to text. While there is limited clarification about the type of transcription (beyond ‘verbatim’), the discussion of the subjective decision-making as part of the transcription process and acknowledgment of the agency of the individuals undertaking transcription increases transparency and therefore rigor.
Conclusions
Qualitative research can help us to understand some of the important issues impacting the third sector in ways that quantitative methods fall short of explaining, such as the ways that individuals and organizations make sense of public policy and societal challenges, how and why organizations design their services and activities in particular ways, and the intricacies of the relationships between boards, executives, staff and volunteers. Qualitative methods training stresses that an interpretivist epistemological position sees knowledge as socially constructed, yet transcription has slipped through the cracks of methodological examination in the process of creating and interpreting meaning.
In this short article, I sought to draw our attention to this important stage of qualitative data collection and analysis and call on third sector researchers to critically reflect upon transcription both in conducting research and in writing about it. I have focused primarily on the transcription of interviews , rather than focus groups or other multi-person events. All of the points raised in my framework transcription apply to these methods of data collection as well; however, there are further issues that need to be taken into consideration regarding focus groups that warrant further attention, such as the issues of power and accuracy of transcription when there are multiple people speaking and interrupting one another. Researchers employing multi-person recordings should therefore devote more time and consideration to transcription. Finally, technology continues to advance in the area of voice recognition, which may save researchers considerable time and/or money in transcription; however, I implore scholars to see transcription through an interpretivist rather than positivist lens, to ensure that the production of written transcripts is not approached as the creation of objective knowledge.
Appendix 1: Articles Reviewed
- Aasland, A., Kropp, S., & Meylakhs, A. Y. (2020). Between Collaboration and Subordination: State and Non-state Actors in Russian Anti-drug Policy. Voluntas , 31 (2), 422–436. https://doi.org/10.1007/s11266-019-00158-9
- Åberg, P., Einarsson, S., & Reuter, M. (2021). Think Tanks: New Organizational Actors in a Changing Swedish Civil Society. Voluntas , 32 (3), 634–648. https://doi.org/10.1007/s11266-019-00174-9
- Afkhami, A., Nasr Isfahani, A., Abzari, M., & Teimouri, H. (2019). Toward a Deep Insight into Employee Participation in Employer-Supported Volunteering in Iranian Organizations: A Grounded Theory. Voluntas , 30 (5), 1036–1053. https://doi.org/10.1007/s11266-019-00141-4
- Anasti, T. (2020). The Strategic Action Field of Sex Work and Sex Trafficking: A Case Study of a Contentious Field in Chicago. Voluntas , 31 (1), 169–183. https://doi.org/10.1007/s11266-019-00161-0
- Appe, S. (2018). Directions in a Post-aid World? South–South Development Cooperation and CSOs in Latin America. Voluntas , 29 (2), 271–283. https://doi.org/10.1007/s11266-017-9838-0
- Arhin, A. A., Kumi, E., & Adam, M.-A. S. (2018). Facing the Bullet? Non-Governmental Organisations’ (NGOs’) Responses to the Changing Aid Landscape in Ghana. Voluntas , 29 (2), 348–360. https://doi.org/10.1007/s11266-018-9966-1
- Arvidson, M. (2018). Change and Tensions in Non-profit Organizations: Beyond the Isomorphism Trajectory. Voluntas , 29 (5), 898–910. https://doi.org/10.1007/s11266-018-0021-z
- Atia, M., & Herrold, C. E. (2018). Governing Through Patronage: The Rise of NGOs and the Fall of Civil Society in Palestine and Morocco. Voluntas , 29 (5), 1044–1054. https://doi.org/10.1007/s11266-018-9953-6
- Ávila, L., & Amorim, M. (2021). Organisational Identity of Social Enterprises: A Taxonomic Approach. Voluntas , 32 (1), 13–27. https://doi.org/10.1007/s11266-020-00264-z
- Baillie Smith, M., Fadel, B., O’Loghlen, A., & Hazeldine, S. (2020). Volunteering Hierarchies in the Global South: Remuneration and Livelihoods. Voluntas . https://doi.org/10.1007/s11266-020-00254-1
- Bandini, F., Gigli, S., & Mariani, L. (2021). Social Enterprises and Public Value: A Multiple-Case Study Assessment. Voluntas , 32 (1), 61–77. https://doi.org/10.1007/s11266-020-00285-8
- Bano, M. (2019). Partnerships and the Good-Governance Agenda: Improving Service Delivery Through State–NGO Collaborations. Voluntas , 30 (6), 1270–1283. https://doi.org/10.1007/s11266-017-9937-y
- Barinaga, E. (2020). Coopted! Mission Drift in a Social Venture Engaged in a Cross-Sectoral Partnership. Voluntas , 31 (2), 437–449. https://doi.org/10.1007/s11266-018-0019-6
- Bayalieva-Jailobaeva, K. (2018). New Donor Strategies: Implications for NGOs in Post-Soviet Kyrgyzstan. Voluntas , 29 (2), 284–295. https://doi.org/10.1007/s11266-017-9878-5
- Beaton, E. E. (2021). No Margin, No Mission: How Practitioners Justify Nonprofit Managerialization. Voluntas , 32 (3), 695–708. https://doi.org/10.1007/s11266-019-00189-2
- Bergfeld, A., Plagmann, C., & Lutz, E. (2021). Know Your Counterparts: The Importance of Wording for Stakeholder Communication in Social Franchise Enterprises. Voluntas , 32 (1), 104–119. https://doi.org/10.1007/s11266-020-00289-4
- Bidet, E., Eum, H., & Ryu, J. (2018). Diversity of Social Enterprise Models in South Korea. Voluntas , 29 (6), 1261–1273. https://doi.org/10.1007/s11266-018-9951-8
- Bies, A., & Kennedy, S. (2019). The State and the State of the Art on Philanthropy in China. Voluntas , 30 (4), 619–633. https://doi.org/10.1007/s11266-019-00142-3
- Bradford, A., Luke, B., & Furneaux, C. (2020). Exploring Accountability in Social Enterprise: Priorities, Practicalities, and Legitimacy. Voluntas , 31 (3), 614–626. https://doi.org/10.1007/s11266-020-00215-8
- Cannon, S. M. (2020). Legitimacy as Property and Process: The Case of an Irish LGBT Organization. Voluntas , 31 (1), 39–55. https://doi.org/10.1007/s11266-019-00091-x
- Carlsen, H. B., Doerr, N., & Toubøl, J. (2020). Inequality in Interaction: Equalising the Helper–Recipient Relationship in the Refugee Solidarity Movement. Voluntas . https://doi.org/10.1007/s11266-020-00268-9
- Chatterjee, D., Subramanian, B., & Hota, P. K. (2020). Professionalization and Hybridization Dynamics of Social Enterprises. Voluntas , 31 (3), 457–471. https://doi.org/10.1007/s11266-020-00217-6
- Chatzichristos, G., & Nagopoulos, N. (2020). Social Entrepreneurship and Institutional Sustainability: Insights from an Embedded Social Enterprise. Voluntas , 31 (3), 484–493. https://doi.org/10.1007/s11266-019-00188-3
- Chaves-Avila, R., & Savall-Morera, T. (2019). The Social Economy in a Context of Austerity Policies: The Tension Between Political Discourse and Implemented Policies in Spain. Voluntas , 30 (3), 487–498. https://doi.org/10.1007/s11266-018-00075-3
- Chewinski, M. (2019). Coordinating Action: NGOs and Grassroots Groups Challenging Canadian Resource Extraction Abroad. Voluntas , 30 (2), 356–368. https://doi.org/10.1007/s11266-018-0023-x
- Clear, A., Paull, M., & Holloway, D. (2018). Nonprofit Advocacy Tactics: Thinking Inside The Box? Voluntas , 29 (4), 857–869. https://doi.org/10.1007/s11266-017-9907-4
- Clerkin, B., & Quinn, M. (2019). Restricted Funding: Restricting Development? Voluntas , 30 (6), 1348–1364. https://doi.org/10.1007/s11266-018-00048-6
- Cookingham Bailey, E. (2020). Advocacy and Service Delivery in the Voluntary Sector: Exploring the History of Voluntary Sector Activities for New Minority and Migrant Groups in East London, 1970s–1990s. Voluntas . https://doi.org/10.1007/s11266-020-00253-2
- Crack, A. M. (2018). The Regulation of International NGOS: Assessing the Effectiveness of the INGO Accountability Charter. Voluntas , 29 (2), 419–429. https://doi.org/10.1007/s11266-017-9866-9
- Crotty, J., & Ljubownikow, S. (2020). Creating Organisational Strength from Operationalising Restrictions: Welfare Non-profit Organisations in the Russian Federation. Voluntas , 31 (6), 1148–1158. https://doi.org/10.1007/s11266-020-00271-0
- Deng, G. (2019). Trends in Overseas Philanthropy by Chinese Foundations. Voluntas , 30 (4), 678–691. https://doi.org/10.1007/s11266-017-9868-7
- Dinh, K., Hong, K. T., Haire, B., & Worth, H. (2021). Historic and Contemporary Influences on HIV Advocacy in Vietnam. Voluntas , 32 (3), 610–620. https://doi.org/10.1007/s11266-020-00220-x
- Dong, Q., Guo, J., & Huang, C.-C. (2019). Nonprofit Alliance in China: Effects of Alliance Process on Goal Achievement. Voluntas , 30 (2), 300–311. https://doi.org/10.1007/s11266-018-9990-1
- Edenfield, A. C., & Andersson, F. O. (2018). Growing Pains: The Transformative Journey from a Nascent to a Formal Not-For-Profit Venture. Voluntas , 29 (5), 1033–1043. https://doi.org/10.1007/s11266-017-9936-z
- Elsayed, Y. (2018). At the Intersection of Social Entrepreneurship and Social Movements: The Case of Egypt and the Arab Spring. Voluntas , 29 (4), 819–831. https://doi.org/10.1007/s11266-017-9943-0
- Eschweiler, J., Svensson, S., Mocca, E., Cartwright, A., & Villadsen Nielsen, L. (2019). The Reciprocity Dimension of Solidarity: Insights from Three European Countries. Voluntas , 30 (3), 549–561. https://doi.org/10.1007/s11266-018-0031-x
- Eynaud, P., Juan, M., & Mourey, D. (2018). Participatory Art as a Social Practice of Commoning to Reinvent the Right to the City. Voluntas , 29 (4), 621–636. https://doi.org/10.1007/s11266-018-0006-y
- Falkenhain, M. (2020). Dividing Lines: Understanding the Creation and Replication of Fragmentations Among NGOs in Hybrid Regimes. Voluntas , 31 (4), 663–673. https://doi.org/10.1007/s11266-019-00185-6
- Fang, Q., Fisher, K. R., & Li, B. (2020). Follower or Challenger? How Chinese Non-governmental Organizations Manage Accountability Requirements from Funders. Voluntas , 31 (4), 722–735. https://doi.org/10.1007/s11266-019-00184-7
- Farid, M., & Li, H. (2021). Reciprocal Engagement and NGO Policy Influence on the Local State in China. Voluntas , 32 (3), 597–609. https://doi.org/10.1007/s11266-020-00288-5
- Fehsenfeld, M., & Levinsen, K. (2019). Taking Care of the Refugees: Exploring Advocacy and Cross-sector Collaboration in Service Provision for Refugees. Voluntas , 30 (2), 422–435. https://doi.org/10.1007/s11266-019-00097-5
- Ferguson, G. (2018). The Social Economy in Bolivia: Indigeneity, Solidarity, and Alternatives to Capitalism. Voluntas , 29 (6), 1233–1243. https://doi.org/10.1007/s11266-018-0013-z
- Fulton, B. R., & Wood, R. L. (2018). Civil Society Organizations and the Enduring Role of Religion in Promoting Democratic Engagement. Voluntas , 29 (5), 1068–1079. https://doi.org/10.1007/s11266-018-9965-2
- Gaby, S. (2020). Reconfiguring Organizational Commitments: Boundary Crossing in Civic Groups. Voluntas , 31 (6), 1121–1133. https://doi.org/10.1007/s11266-020-00272-z
- Gaeta, G. L., Ghinoi, S., Silvestri, F., & Trasciani, G. (2021). Exploring Networking of Third Sector Organizations: A Case Study Based on the Quartieri Spagnoli Neighborhood in Naples (Italy). Voluntas , 32 (4), 750–766. https://doi.org/10.1007/s11266-020-00241-6
- García-Orosa, B., & Pérez-Seijo, S. (2020). The Use of 360° Video by International Humanitarian Aid Organizations to Spread Social Messages and Increase Engagement. Voluntas , 31 (6), 1311–1329. https://doi.org/10.1007/s11266-020-00280-z
- Grazioli, M., & Caciagli, C. (2018). Resisting to the Neoliberal Urban Fabric: Housing Rights Movements and the Re-appropriation of the ‘Right to the City’ in Rome, Italy. Voluntas , 29 (4), 697–711. https://doi.org/10.1007/s11266-018-9977-y
- Grubb, A., & Henriksen, L. S. (2019). On the Changing Civic Landscape in Denmark and its Consequences for Civic Action. Voluntas , 30 (1), 62–73. https://doi.org/10.1007/s11266-018-00054-8
- Guha, P. (2019). Going to Scale: A Case Study of an Indian Educational NGO. Voluntas , 30 (6), 1365–1379. https://doi.org/10.1007/s11266-019-00099-3
- Guo, C., & Lai, W. (2019). Community Foundations in China: In Search of Identity? Voluntas , 30 (4), 647–663. https://doi.org/10.1007/s11266-017-9932-3
- Heckert, R., Boumans, J., & Vliegenthart, R. (2020). How to Nail the Multiple Identities of an Organization? A Content Analysis of Projected Identity. Voluntas , 31 (1), 129–141. https://doi.org/10.1007/s11266-019-00157-w
- Heideman, L. J. (2018). Making Civil Society Sustainable: The Legacy of USAID in Croatia. Voluntas , 29 (2), 333–347. https://doi.org/10.1007/s11266-017-9896-3
- Hjort, M., & Beswick, D. (2020). Volunteering and Policy Makers: The Political Uses of the UK Conservative Party’s International Development Volunteering Projects. Voluntas . https://doi.org/10.1007/s11266-020-00222-9
- Holthaus, L. (2019). Furthering Pluralism? The German Foundations in Transitional Tunisia. Voluntas , 30 (6), 1284–1296. https://doi.org/10.1007/s11266-018-00074-4
- Horvath, A. (2020). The Transformative Potential of Experience: Learning, Group Dynamics, and the Development of Civic Virtue in a Mobile Soup Kitchen. Voluntas , 31 (5), 981–994. https://doi.org/10.1007/s11266-018-00062-8
- Horvath, A., Brandtner, C., & Powell, W. W. (2018). Serve or Conserve: Mission, Strategy, and Multi-Level Nonprofit Change During the Great Recession. Voluntas , 29 (5), 976–993. https://doi.org/10.1007/s11266-017-9948-8
- Huang, Y. (2019). At the Mercy of the State: Health Philanthropy in China. Voluntas , 30 (4), 634–646. https://doi.org/10.1007/s11266-018-9960-7
- Ismail, A., & Johnson, B. (2019). Managing Organizational Paradoxes in Social Enterprises: Case Studies from the MENA Region. Voluntas , 30 (3), 516–534. https://doi.org/10.1007/s11266-018-00083-3
- Jezierska, K., & Polanska, D. V. (2018). Social Movements Seen as Radical Political Actors: The Case of the Polish Tenants’ Movement. Voluntas , 29 (4), 683–696. https://doi.org/10.1007/s11266-017-9917-2
- Kampen, T., Veldboer, L., & Kleinhans, R. (2019). The Obligation to Volunteer as Fair Reciprocity? Welfare Recipients’ Perceptions of Giving Back to Society. Voluntas , 30 (5), 991–1005. https://doi.org/10.1007/s11266-018-00082-4
- Kang, Y. (2019). What Does China’s Twin-Pillared NGO Funding Game Entail? Growing Diversity and Increasing Isomorphism. Voluntas , 30 (3), 499–515. https://doi.org/10.1007/s11266-018-00085-1
- Kappelides, P., Cuskelly, G., & Hoye, R. (2019). The Influence of Volunteer Recruitment Practices and Expectations on the Development of Volunteers’ Psychological Contracts. Voluntas , 30 (1), 259–271. https://doi.org/10.1007/s11266-018-9986-x
- Kennedy, D. (2019). The Inherently Contested Nature of Nongovernmental Accountability: The Case of HAP International. Voluntas , 30 (6), 1393–1405. https://doi.org/10.1007/s11266-019-00134-3
- Kewes, A., & Munsch, C. (2019). Should I Stay or Should I Go? Engaging and Disengaging Experiences in Welfare-Sector Volunteering. Voluntas , 30 (5), 1090–1103. https://doi.org/10.1007/s11266-019-00122-7
- Korstenbroek, T., & Smets, P. (2019). Developing the Potential for Change: Challenging Power Through Social Entrepreneurship in the Netherlands. Voluntas , 30 (3), 475–486. https://doi.org/10.1007/s11266-019-00107-6
- Koubek, M. (2020). When Did They Protest? Beyond Co-optation or Channeling: Effects of EU Funding on Czech Romani NGOs and Civil Society. Voluntas , 31 (2), 404–421. https://doi.org/10.1007/s11266-018-0008-9
- Kravchenko, Z., & Moskvina, A. (2018). Entrepreneurial NPOs in Russia: Rationalizing the Mission. Voluntas , 29 (5), 962–975. https://doi.org/10.1007/s11266-018-0016-9
- Kumi, E. (2019). Aid Reduction and NGDOs’ Quest for Sustainability in Ghana: Can Philanthropic Institutions Serve as Alternative Resource Mobilisation Routes? Voluntas , 30 (6), 1332–1347. https://doi.org/10.1007/s11266-017-9931-4
- Lall, S. A. (2019). From Legitimacy to Learning: How Impact Measurement Perceptions and Practices Evolve in Social Enterprise–Social Finance Organization Relationships. Voluntas , 30 (3), 562–577. https://doi.org/10.1007/s11266-018-00081-5
- Lang, R., & Mullins, D. (2020). Field Emergence in Civil Society: A Theoretical Framework and Its Application to Community-Led Housing Organisations in England. Voluntas , 31 (1), 184–200. https://doi.org/10.1007/s11266-019-00138-z
- Laurent, A., Garaudel, P., Schmidt, G., & Eynaud, P. (2020). Civil Society Meta-organizations and Legitimating Processes: The Case of the Addiction Field in France. Voluntas , 31 (1), 19–38. https://doi.org/10.1007/s11266-019-00094-8
- Lee, E. K. M., & Chandra, Y. (2020). Dynamic and Marketing Capabilities as Predictors of Social Enterprises’ Performance. Voluntas , 31 (3), 587–600. https://doi.org/10.1007/s11266-019-00155-y
- Li, S. (2020). Global Civil Society Under the New INGO Regulatory Law: A Comparative Case Study on Two INGOs in China. Voluntas , 31 (4), 751–761. https://doi.org/10.1007/s11266-019-00101-y
- Li, S., & Wang, X. (2020). Seeking Credibility from Uncertainty: How Formal Cooptation Institution Unleashes Outspoken NGOs. Voluntas , 31 (4), 711–721. https://doi.org/10.1007/s11266-020-00204-x
- Liu, F., & Zhang, X. (2020). Who are Volunteers in Urban China? Voluntas . https://doi.org/10.1007/s11266-020-00251-4
- Lund, V., & Juujärvi, S. (2018). Residents’ Agency Makes a Difference in Volunteering in an Urban Neighbourhood. Voluntas , 29 (4), 756–769. https://doi.org/10.1007/s11266-018-9955-4
- Marie, B., Isabelle, M., Jordan, B.-L., Hélène, C., Sophie, É., Julie, F., Christine, M., & Andrée, S. (2020). Promising Practices of Nonprofit Organizations to Respond to the Challenges Faced in Countering the Mistreatment of Older Adults. Voluntas , 31 (6), 1359–1370. https://doi.org/10.1007/s11266-020-00252-3
- Martinez, D. E., & Cooper, D. J. (2020). Seeing Through the Logical Framework. Voluntas , 31 (6), 1239–1253. https://doi.org/10.1007/s11266-020-00223-8
- Mathews, M. A. (2020). The Embeddedness of Nonprofit Leadership in Civic Governance. Voluntas , 31 (1), 201–212. https://doi.org/10.1007/s11266-019-00139-y
- Mati, J. M. (2020). Civil Society in ‘Politics’ and ‘Development’ in African Hybrid Regimes: The Kenyan Case. Voluntas , 31 (4), 674–687. https://doi.org/10.1007/s11266-020-00211-y
- Maya-Jariego, I., Holgado-Ramos, D., González-Tinoco, E., Muñoz-Alvis, A., & Ortega, M. (2020). More Money, More Problems? Resource Dependence and Professionalization of Non-governmental Social Services Organizations in Southern Spain. Voluntas , 31 (6), 1212–1225. https://doi.org/10.1007/s11266-020-00256-z
- McLennan, B. J. (2020). Conditions for Effective Coproduction in Community-Led Disaster Risk Management. Voluntas , 31 (2), 316–332. https://doi.org/10.1007/s11266-018-9957-2
- McMullin, C., & Skelcher, C. (2018). The Impact of Societal-Level Institutional Logics on Hybridity: Evidence from Nonprofit Organizations in England and France. Voluntas , 29 (5), 911–924. https://link.springer.com/article/10.1007/s11266-018-9996-8
- Meijs, L., Handy, F., Simons, F.-J., & Roza, L. (2020). A Social Innovation: Addressing Relative Food Insecurity and Social Exclusion. Voluntas , 31 (5), 894–906. https://doi.org/10.1007/s11266-019-00105-8
- Meyer, M., & Simsa, R. (2018). Organizing the Unexpected: How Civil Society Organizations Dealt with the Refugee Crisis. Voluntas , 29 (6), 1159–1175. https://doi.org/10.1007/s11266-018-00050-y
- Mikołajczak, P. (2020). Social Enterprises’ Hybridity in the Concept of Institutional Logics: Evidence from Polish NGOs. Voluntas , 31 (3), 472–483. https://doi.org/10.1007/s11266-020-00195-9
- Milbourn, B., Black, M. H., & Buchanan, A. (2019). Why People Leave Community Service Organizations: A Mixed Methods Study. Voluntas , 30 (1), 272–281. https://doi.org/10.1007/s11266-018-0005-z
- Molina, J. L., Valenzuela-García, H., Lubbers, M. J., Escribano, P., & Lobato, M. M. (2018). “The Cowl Does Make The Monk”: Understanding the Emergence of Social Entrepreneurship in Times of Downturn. Voluntas , 29 (4), 725–739. https://doi.org/10.1007/s11266-017-9921-6
- Moraes, R. L., & Andion, C. (2018). Civil Society and Social Innovation in Public Arenas in Brazil: Trajectory and Experience of the Movement Against Electoral Corruption (MCCE). Voluntas , 29 (4), 801–818. https://doi.org/10.1007/s11266-017-9867-8
- Morais-da-Silva, R. L., Segatto, A. P., & Bezerra-de-Sousa, I. G. (2020). Connecting Two Sides: A Qualitative Study on Social Innovation Ventures and Poor Communities in an Emerging Economy. Voluntas , 31 (5), 966–980. https://doi.org/10.1007/s11266-019-00156-x
- Musah-Surugu, I. J., Bawole, J. N., & Ahenkan, A. (2019). The “Third Sector” and Climate Change Adaptation Governance in Sub-Saharan Africa: Experience from Ghana. Voluntas , 30 (2), 312–326. https://doi.org/10.1007/s11266-018-9962-5
- Mutongwizo, T. (2018). Comparing NGO Resilience and ‘Structures of Opportunity’ in South Africa and Zimbabwe (2010–2013). Voluntas , 29 (2), 373–387. https://doi.org/10.1007/s11266-017-9881-x
- Näslund, H. (2020). Collective Deliberations and Hearts on Fire: Experiential Knowledge Among Entrepreneurs and Organisations in the Mental Health Service User Movement. Voluntas . https://doi.org/10.1007/s11266-020-00233-6
- Nimu, A. (2018). Surviving Mechanisms and Strategies of Gender Equality NGOs in Romania and Poland. Voluntas , 29 (2), 310–332. https://doi.org/10.1007/s11266-018-9961-6
- Noh, J.-E. (2019). Human Rights-Based Child Sponsorship: A Case Study of ActionAid. Voluntas , 30 (6), 1420–1432. https://doi.org/10.1007/s11266-018-0010-2
- Obaze, Y. (2020). Supply Chain Challenges and Shared Value Destruction in the Community-Based Supply Chain. Voluntas , 31 (3), 550–562. https://doi.org/10.1007/s11266-020-00202-z
- Onyx, J., Darcy, S., Grabowski, S., Green, J., & Maxwell, H. (2018). Researching the Social Impact of Arts and Disability: Applying a New Empirical Tool and Method. Voluntas , 29 (3), 574–589. https://doi.org/10.1007/s11266-018-9968-z
- Pape, U., Brandsen, T., Pahl, J. B., Pieliński, B., Baturina, D., Brookes, N., Chaves-Ávila, R., Kendall, J., Matančević, J., Petrella, F., Rentzsch, C., Richez-Battesti, N., Savall-Morera, T., Simsa, R., & Zimmer, A. (2020). Changing Policy Environments in Europe and the Resilience of the Third Sector. Voluntas , 31 (1), 238–249. https://doi.org/10.1007/s11266-018-00087-z
- Parker, M. A., Mook, L., Kao, C.-Y., & Murdock, A. (2020). Accountability and Relationship-Definition Among Food Banks Partnerships. Voluntas , 31 (5), 923–937. https://doi.org/10.1007/s11266-019-00150-3
- Peci, A., Oquendo, M. I., & Mendonça, P. (2020). Collaboration, (Dis)trust and Control in Brazilian Manufactured Public/Non-profit Partnerships. Voluntas , 31 (2), 375–389. https://doi.org/10.1007/s11266-018-0027-6
- Peralta, K. J., & Vaitkus, E. (2019). Constructing Action: An Analysis of the Roles of Third Sector Actors During the Implementation of the Dominican Republic’s Regularization Plan. Voluntas , 30 (6), 1319–1331. https://doi.org/10.1007/s11266-018-0003-1
- Pettigrew, S., Jongenelis, M., Jackson, B., & Newton, R. U. (2019). “Charity Begins at Home”: Informal Caring Barriers to Formal Volunteering Among Older People. Voluntas , 30 (5), 921–931. https://doi.org/10.1007/s11266-018-0017-8
- Pfeilstetter, R. (2020). Doing Good and Selling Goods. Voluntas , 31 (3), 511–520. https://doi.org/10.1007/s11266-020-00194-w
- Pixová, M. (2018). The Empowering Potential of Reformist Urban Activism in Czech Cities. Voluntas , 29 (4), 670–682. https://doi.org/10.1007/s11266-018-0011-1
- Pluciński, P. (2018). Forces of Altermodernization: Urban Social Movements and the New Urban Question in Contemporary Poland. Voluntas , 29 (4), 653–669. https://doi.org/10.1007/s11266-018-0007-x
- Popplewell, R. (2018). Civil Society, Legitimacy and Political Space: Why Some Organisations are More Vulnerable to Restrictions than Others in Violent and Divided Contexts. Voluntas , 29 (2), 388–403. https://doi.org/10.1007/s11266-018-9949-2
- Puljek-Shank, R. (2018). Civic Agency in Governance: The Role of Legitimacy with Citizens vs. Donors. Voluntas , 29 (4), 870–883. https://doi.org/10.1007/s11266-018-0020-0
- Reijnders, M., Schalk, J., & Steen, T. (2018). Services Wanted? Understanding the Non-take-up of Social Support at the Local Level. Voluntas , 29 (6), 1360–1374. https://doi.org/10.1007/s11266-018-00060-w
- Ruiz Sportmann, A. S., & Greenspan, I. (2019). Relational Interactions Between Immigrant and Native-Born Volunteers: Trust-Building and Integration or Suspicion and Conflict? Voluntas , 30 (5), 932–946. https://doi.org/10.1007/s11266-019-00108-5
- Sandberg, B., Elliott, E., & Petchel, S. (2020). Investigating the Marketization of the Nonprofit Sector: A Comparative Case Study of Two Nonprofit Organizations. Voluntas , 31 (3), 494–510. https://doi.org/10.1007/s11266-019-00159-8
- Schmid, H., & Almog-Bar, M. (2020). The Critical Role of the Initial Stages of Cross-Sector Partnerships and Their Implications for Partnerships’ Outcomes. Voluntas , 31 (2), 286–300. https://doi.org/10.1007/s11266-019-00137-0
- Sepulveda, L., Lyon, F., & Vickers, I. (2020). Implementing Democratic Governance and Ownership: The Interplay of Structure and Culture in Public Service Social Enterprises. Voluntas , 31 (3), 627–641. https://doi.org/10.1007/s11266-020-00201-0
- Shier, M. L., & Handy, F. (2020). Leadership in Nonprofits: Social Innovations and Blurring Boundaries. Voluntas , 31 (2), 333–344. https://doi.org/10.1007/s11266-018-00078-0
- Spencer, S. B., & Skalaban, I. A. (2018). Organizational Culture in Civic Associations in Russia. Voluntas , 29 (5), 1080–1097. https://doi.org/10.1007/s11266-017-9925-2
- Stewart, A. J., & Twumasi, A. (2020). Minding the Gap: An Exploratory Study Applying Theory to Nonprofit Board Management of Executive Transitions. Voluntas , 31 (6), 1268–1281. https://doi.org/10.1007/s11266-020-00244-3
- Stougaard, M. S. (2020). Co-producing Public Welfare Services with Vulnerable Citizens: A Case Study of a Danish-Somali Women’s Association Co-producing Crime Prevention with the Local Authorities. Voluntas . https://doi.org/10.1007/s11266-020-00235-4
- Strichman, N., Marshood, F., & Eytan, D. (2018). Exploring the Adaptive Capacities of Shared Jewish–Arab Organizations in Israel. Voluntas , 29 (5), 1055–1067. https://doi.org/10.1007/s11266-017-9904-7
- Suter, P., & Gmür, M. (2018). Volunteer Engagement in Housing Co-operatives: Civil Society “en miniature”. Voluntas , 29 (4), 770–789. https://doi.org/10.1007/s11266-018-9959-0
- Svensson, K., & Gallo, C. (2018). The Creation of an Unsolicited Organization: Victim Support Sweden. Voluntas , 29 (5), 1008–1018. https://doi.org/10.1007/s11266-018-9978-x
- Tadesse, H. A., & Steen, T. (2019). Exploring the Impact of Political Context on State–Civil Society Relations: Actors’ Strategies in a Developmental State. Voluntas , 30 (6), 1256–1269. https://doi.org/10.1007/s11266-018-00077-1
- Taylor, R., Torugsa, N. (Ann), & Arundel, A. (2020). Organizational Pathways for Social Innovation and Societal Impacts in Disability Nonprofits. Voluntas , 31 (5), 995–1012. https://doi.org/10.1007/s11266-019-00113-8
- Uzcanga, C., & Oiarzabal, P. J. (2019). Associations of Migrants in Spain: An Enquiry into Their Digital Inclusion in the “Network Society” in the 2010s. Voluntas , 30 (5), 947–961. https://doi.org/10.1007/s11266-019-00162-z
- van der Veer, L. (2020). Treacherous Elasticity, Callous Boundaries: Aspiring Volunteer Initiatives in the Field of Refugee Support in Rotterdam. Voluntas . https://doi.org/10.1007/s11266-020-00260-3
- van Wessel, M., Naz, F., & Sahoo, S. (2021). Complementarities in CSO Collaborations: How Working with Diversity Produces Advantages. Voluntas , 32 (4), 717–730. https://doi.org/10.1007/s11266-020-00227-4
- Vandepitte, E., Vandermoere, F., & Hustinx, L. (2019). Civil Anarchizing for the Common Good: Culturally Patterned Politics of Legitimacy in the Climate Justice Movement. Voluntas , 30 (2), 327–341. https://doi.org/10.1007/s11266-018-00073-5
- Vuković, D. (2018). The Quest for Government Accountability and Rule of Law: Conflicting Strategies of State and Civil Society in Cambodia and Serbia. Voluntas , 29 (3), 590–602. https://doi.org/10.1007/s11266-018-9984-z
- Waardenburg, M. (2021). Understanding the Microfoundations of Government–Civil Society Relations. Voluntas , 32 (3), 548–560. https://doi.org/10.1007/s11266-020-00221-w
- Wells, R., & Anasti, T. (2020). Hybrid Models for Social Change: Legitimacy Among Community-Based Nonprofit Organizations. Voluntas , 31 (6), 1134–1147. https://doi.org/10.1007/s11266-019-00126-3
- Williamson, A. K., Luke, B., & Furneaux, C. (2021). Ties That Bind: Public Foundations in Dyadic Partnerships. Voluntas , 32 (2), 234–246. https://doi.org/10.1007/s11266-020-00269-8
- Yanay-Ventura, G. (2019). “Nothing About Us Without Us” in Volunteerism Too: Volunteering Among People with Disabilities. Voluntas , 30 (1), 147–163. https://doi.org/10.1007/s11266-018-0026-7
- Yanay-Ventura, G., Issaq, L., & Sharabi, M. (2020). Civic Service and Social Class: The Case of Young Arab Women in Israel. Voluntas . https://doi.org/10.1007/s11266-020-00210-z
- Zhang, Y., Bradtke, M., & Halvey, M. (2020). Anxiety and Ambivalence: NGO–Activist Partnership in China’s Environmental Protests, 2007–2016. Voluntas , 31 (4), 779–792. https://doi.org/10.1007/s11266-020-00237-2
- Zihnioğlu, Ö. (2019). The Prospects of Civic Alliance: New Civic Activists Acting Together with Civil Society Organizations. Voluntas , 30 (2), 289–299. https://doi.org/10.1007/s11266-018-0032-9
No funding was received to assist with the preparation of this manuscript.
Declarations
The author declares that they have no conflict of interest.
1 While this approach may have obscured other methods that employ transcription, such as focus groups, the intention of the survey is to provide a snapshot illustration of transcription and qualitative methods rather than a systematic review.
2 Articles reviewed are listed in Appendix 1.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
- Atia M, Herrold CE. Governing through patronage: The rise of NGOs and the fall of civil society in Palestine and Morocco. VOLUNTAS: International Journal of Voluntary and Nonprofit Organizations. 2018; 29 (5):1044–1054. doi: 10.1007/s11266-018-9953-6. [ CrossRef ] [ Google Scholar ]
- Blaxter, L., Hughes, C., & Tight, M. (2001). How to research (2nd ed.). Open University Press.
- Bokhove C, Downey C. Automated generation of “good enough” transcripts as a first step to transcription of audio-recorded data. Methodological Innovations. 2018; 11 (2):2059799118790743. doi: 10.1177/2059799118790743. [ CrossRef ] [ Google Scholar ]
- Braun V, Clarke V. Using thematic analysis in psychology. Qualitative Research in Psychology. 2006; 3 (2):77–101. doi: 10.1191/1478088706qp063oa. [ CrossRef ] [ Google Scholar ]
- Bryman, A. (2016). Social research methods (5th ed.). Oxford University Press.
- Bucholtz M. The politics of transcription. Journal of Pragmatics. 2000; 32 (10):1439–1465. doi: 10.1016/S0378-2166(99)00094-6. [ CrossRef ] [ Google Scholar ]
- Da Silva J. Producing ‘good enough’ automated transcripts securely: Extending Bokhove and Downey (2018) to address security concerns. Methodological Innovations. 2021; 14 (1):2059799120987766. doi: 10.1177/2059799120987766. [ CrossRef ] [ Google Scholar ]
- Davidson C. Transcription: Imperatives for qualitative research. International Journal of Qualitative Methods. 2009; 8 (2):35–52. doi: 10.1177/160940690900800206. [ CrossRef ] [ Google Scholar ]
- Dodds S, Hess AC. Adapting research methodology during COVID-19: Lessons for transformative service research. Journal of Service Management. 2020; 32 (2):203–217. doi: 10.1108/JOSM-05-2020-0153. [ CrossRef ] [ Google Scholar ]
- Duranti A. Transcripts, like shadows on a wall. Mind, Culture, and Activity. 2006; 13 (4):301–310. doi: 10.1207/s15327884mca1304_3. [ CrossRef ] [ Google Scholar ]
- Gibbs, G. R., Friese, S., & Mangabeira, W. C. (2002). View of the use of new technology in qualitative research. Orum Qualitative Sozialforschung/Forum: Qualitative Social Research , 3 (2). https://www.qualitative-research.net/index.php/fqs/article/view/847/1840
- Halcomb EJ, Davidson PM. Is verbatim transcription of interview data always necessary? Applied Nursing Research. 2006; 19 (1):38–42. doi: 10.1016/j.apnr.2005.06.001. [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Igalla M, Edelenbos J, van Meerkerk I. Citizens in action, what do they accomplish? A systematic literature review of citizen initiatives, their main characteristics, outcomes, and factors. VOLUNTAS: International Journal of Voluntary and Nonprofit Organizations. 2019; 30 (5):1176–1194. doi: 10.1007/s11266-019-00129-0. [ CrossRef ] [ Google Scholar ]
- Jaffe A. Variability in transcription and the complexities of representation, authority and voice. Discourse Studies. 2007; 9 (6):831–836. doi: 10.1177/1461445607082584. [ CrossRef ] [ Google Scholar ]
- Kvale, S. (1996). Interviews: An introduction to qualitative research interviewing . Sage.
- Lapadat JC. Problematizing transcription: Purpose, paradigm and quality. International Journal of Social Research Methodology. 2000; 3 (3):203–219. doi: 10.1080/13645570050083698. [ CrossRef ] [ Google Scholar ]
- Laurett R, Ferreira JJ. Strategy in nonprofit organisations: A systematic literature review and agenda for future research. VOLUNTAS: International Journal of Voluntary and Nonprofit Organizations. 2018; 29 (5):881–897. doi: 10.1007/s11266-017-9933-2. [ CrossRef ] [ Google Scholar ]
- Loubere, N. (2017). Questioning transcription: The case for the systematic and reflexive interviewing and reporting (SRIR) method. Forum Qualitative Sozialforschung/Forum: Qualitative Social Research , 18 (2), Article 2. 10.17169/fqs-18.2.2739
- Markle, D. T., West, R. E., & Rich, P. J. (2011). Beyond transcription: Technology, change, and refinement of method. Forum Qualitative Sozialforschung/Forum: Qualitative Social Research , 12 (3), Article 3. 10.17169/fqs-12.3.1564
- Matheson JL. The voice transcription technique: Use of voice recognition software to transcribe digital interview data in qualitative research. Qualitative Report. 2007; 12 (4):547–560. [ Google Scholar ]
- McCorkel JA, Myers K. What difference does difference make? Position and privilege in the field. Qualitative Sociology. 2003; 26 (2):199–231. doi: 10.1023/A:1022967012774. [ CrossRef ] [ Google Scholar ]
- Mero-Jaffe I. ‘Is that what I Said?’ Interview transcript approval by participants: An aspect of ethics in qualitative research. International Journal of Qualitative Methods. 2011; 10 (3):231–247. doi: 10.1177/160940691101000304. [ CrossRef ] [ Google Scholar ]
- Mishler, E. (2003). Representing discourse: The rhetoric of transcription. In N. Fielding (Ed.), Interviewing . Sage.
- Moravcsik, A. (2014). Transparency: The revolution in qualitative research. PS: Political Science & Politics , 47 (1), 48–53. 10.1017/S1049096513001789
- Oliver DG, Serovich JM, Mason TL. Constraints and opportunities with interview transcription: Towards reflection in qualitative research. Social Forces. 2005; 84 (2):1273–1289. doi: 10.1353/sof.2006.0023. [ PMC free article ] [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Packer MJ. The science of qualitative research. Cambridge University Press; 2017. [ Google Scholar ]
- Parameswaran UD, Ozawa-Kirk JL, Latendresse G. To live (code) or to not: A new method for coding in qualitative research. Qualitative Social Work. 2020; 19 (4):630–644. doi: 10.1177/1473325019840394. [ CrossRef ] [ Google Scholar ]
- Quintanilha M, Mayan MJ, Thompson J, Bell RC. Different approaches to cross-lingual focus groups: Lessons from a cross-cultural community-based participatory research project in the ENRICH study. International Journal of Qualitative Methods. 2015; 14 (5):1609406915621419. doi: 10.1177/1609406915621419. [ CrossRef ] [ Google Scholar ]
- Schwingel, A., Wiley, A., Teran-Garcia, M., McCaffrey, J., Gálvez, P., Hawn, R., Villegas, E., Coba, S., Vizcarra, M., Luty, G., Montez, R., & The Abriendo Caminos Promotora Project Group. (2017). More than help? Volunteerism in US Latino culture. VOLUNTAS: International Journal of Voluntary and Nonprofit Organizations , 28 (1), 162–183. 10.1007/s11266-016-9731-2
- Stelma JH, Cameron LJ. Intonation units in spoken interaction: Developing transcription skills. Text & Talk. 2007; 27 (3):361–393. doi: 10.1515/TEXT.2007.015. [ CrossRef ] [ Google Scholar ]
- Tilley SA. “Challenging” research practices: Turning a critical lens on the work of transcription. Qualitative Inquiry. 2003; 9 (5):750–773. doi: 10.1177/1077800403255296. [ CrossRef ] [ Google Scholar ]
- Tracy, S. J. (2019). Qualitative research methods: Collecting evidence, crafting analysis (2nd ed.). John Wiley & Sons.
- von Schnurbein G, Perez M, Gehringer T. Nonprofit comparative research: Recent agendas and future trends. VOLUNTAS: International Journal of Voluntary and Nonprofit Organizations. 2018; 29 (3):437–453. doi: 10.1007/s11266-017-9877-6. [ CrossRef ] [ Google Scholar ]
- Wellard S, McKenna L. Turning tapes into text: Issues surrounding the transcription of interviews. Contemporary Nurse. 2001; 11 (2–3):180–186. doi: 10.5172/conu.11.2-3.180. [ PubMed ] [ CrossRef ] [ Google Scholar ]
- Yanay-Ventura, G., Issaq, L., & Sharabi, M. (2020). Civic service and social class: The case of young Arab women in Israel. VOLUNTAS: International Journal of Voluntary and Nonprofit Organizations . 10.1007/s11266-020-00210-z
- Correspondence
- Open access
- Published: 18 June 2024
The CDK9 inhibitor enitociclib overcomes resistance to BTK inhibition and CAR-T therapy in mantle cell lymphoma
- Vivian Jiang 1 ,
- William Lee 1 ,
- Tianci Zhang 1 ,
- Alexa Jordan 1 ,
- Fangfang Yan 1 ,
- Qingsong Cai 1 ,
- Joseph McIntosh 1 ,
- Jovanny Vargas 1 ,
- Yang Liu 1 &
- Michael Wang 1 , 2
Biomarker Research volume 12 , Article number: 62 ( 2024 ) Cite this article
84 Accesses
Metrics details
Inhibitors of Bruton’s tyrosine kinase (BTKi) and chimeric antigen receptor T-cell (CAR-T) therapy targeting CD19 are paradigm-shifting advances in treating patients with aggressive mantle cell lymphoma (MCL). However, clinical relapses following BTKi and CD19-directed CAR-T treatments are a fast-growing medical challenge. Development of novel therapies to overcome BTKi resistance (BTKi-R) and BTKi-CAR-T dual resistance (Dual-R) are urgently needed. Our single-cell RNA sequencing data revealed major transcriptomic reprogramming, with great enrichment of MYC-targets evolving as resistance to these therapies developed. Interestingly, cyclin-dependent kinase 9 (CDK9), a critical component of the positive transcription elongation factor-b complex, was among the top upregulated genes in Dual-R vs. BTKi-R samples. We therefore hypothesized that targeting CDK9 may turn off MYC-driven tumor survival and drug resistance. Enitociclib (formerly VIP152) is a selective CDK9 inhibitor whose potency against MCL has not been assessed. In this study, we found that enitociclib was highly potent in targeting lymphoma cells, with the half-maximal inhibitory concentration (IC 50 ) ranging from 32 to 172 nM in MCL and diffuse large B-cell lymphoma cell lines. It inhibited CDK9 phosphorylation and downstream events including de novo synthesis of the short-lived proteins c-MYC, MCL-1, and cyclin D1, and induced apoptosis in a caspase-3-dependent manner. Enitociclib potently inhibited in vivo tumor growth of cell line-derived and patient-derived xenografts having therapeutic resistance. Our data demonstrate the potency of enitociclib in overcoming therapeutic resistance in MCL models and provide evidence in favor of its clinical investigation.
To the editor
Mantle cell lymphoma [ 1 ] is a very aggressive subtype of non-Hodgkin lymphoma. There have been paradigm-shifting therapeutic advances in the last decade, including BTKi therapies (ibrutinib, acalabrutinib, zanubrutinib, and pirtobrutinib) and anti-CD19 CAR-T therapy [ 2 , 3 , 4 , 5 , 6 ]. However, therapeutic relapse frequently occurs, and there is a rising need to prevent or overcome resistance in patients who relapse. Our single-cell RNA sequencing data showed that MYC targets were progressively enriched with BTKi resistance (Fig. 1 A). MYC mRNA expression was upregulated in BTKi-R compared to BTKi-sensitive (BTKi-S) cells, and its high expression correlated with poor patient survival in our patient cohort ( p = 0.037) (Fig. 1 B) and two independent cohorts [ 7 , 8 ] ( p = 0.0032 and 0.0027, respectively) (Supplementary Figure S1 A-B). Moreover, cyclin-dependent kinase 9 (CDK9) was among the top upregulated genes in Dual-R samples vs. solely BTKi-R samples [ 9 ]. Therefore, we targeted the transcription gatekeeper CDK9 to see if that approach could overcome therapeutic resistance. CDK9 inhibition by small molecules such as AZD4573 induces acute loss of short-lived mRNA and proteins, including c-MYC and MCL-1 [ 10 ]. Enitociclib is a selective and potent CDK9 inhibitor with a better safety profile than AZD4573 [ 11 ]; however, its potency in treating MCL and whether it overcomes therapeutic resistance is not known.
Enitociclib was highly potent in primary MCL cells, MCL cell lines, and diffuse large B-cell lymphoma (DLBCL) cell lines, with an IC 50 of 32–172 nM (Fig. 1 C and Supplementary Figure S2 A). Enitociclib inhibited cell viability in a dose- and time-dependent manner by robustly inducing apoptosis (Fig. 1D and Supplementary Figure S2 B-C and S3 A-B). Upon treatment with enitociclib for 6 h, CDK9 phosphorylation was markedly inhibited in both JeKo-R cells with acquired BTKi-resistance and Z138 cells with primary BTKi-resistance (Fig. 1 E). Correspondingly, CDK9 downstream signaling events, including phosphorylation of RNA polymerase II (Pol II) at Ser 2, were also downregulated, along with reduced expression of the short-lived proteins c-MYC, MCL-1, and cyclin D1 (Fig. 1 E). These changes were dose-dependent in JeKo-R cells (Fig. 1 F). Similarly, cycloheximide (CHX), a translation elongation inhibitor, diminished c-MYC expression, while the proteasome inhibitor MG132 failed to rescue enitociclib-induced c-MYC downregulation (Fig. 1 G and Supplementary Figure S4 ). These indicate that enitociclib blocks de novo gene expression of short-lived proteins but not protein degradation.
Additionally, the two apoptosis indicators, cleaved PARP (poly (ADP-ribose) polymerase) and cleaved caspase-3, were markedly upregulated upon CDK9 inhibition (Fig. 1 E). Enitociclib treatment triggered apoptosis as early as 6 h and further augmented it at 24 and 48 h in JeKo-R cells (Fig. 1 H). Enitociclib-triggered apoptosis was blocked by the pan-caspase inhibitor Z-VAD-FMK and by the specific caspase-3 inhibitor Z-VEAD-FMK in JeKo-R and JeKo-1 cells (Fig. 1 I-L and Supplementary Figure S5 A-B). Together, these data demonstrate that targeting CDK9 with enitociclib triggered apoptosis in a caspase-3-dependent manner.
To determine whether targeting CDK9 with enitociclib can effectively and safely overcome therapeutic resistance in MCL, we first tested its in vivo efficacy using JeKo-1 cell line-derived xenografts (CDXs). Enitociclib at 10 mg/kg (IV, twice a week) markedly inhibited the tumor growth of JeKo-1 CDXs in immunodeficient NSG (NOD.Cg-Prkdc scid Il2rg tm1Wjl /SzJ) mice ( p < 0.0001) and correspondingly prolonged mouse survival ( p < 0.0001) (Fig. 2 A-B) without significant body weight loss (Fig. 2 C) or other apparent adverse effect. To further address this, we established patient-derived xenograft (PDX) models from three patients with different types of therapeutic resistance: PDX-1 having BTKi resistance (Fig. 2 D-F), PDX-2 having dual resistance to the BTKi ibrutinib and the Bcl-2 inhibitor venetoclax (Fig. 2 G-I), and PDX-3 having dual BTKi-CAR-T resistance (Fig. 2 J-L). Enitociclib efficaciously inhibited in vivo PDX growth of PDX-1 ( p = 0.00015), PDX-2 ( p = 0.009), and PDX-3 ( p = 0.000003) without causing significant body weight loss (Fig. 2 D-L).
Our findings showed that targeting CDK9 with its specific inhibitor enitociclib led to potent anti-lymphoma activity in vitro and in vivo. Enitociclib induced rapid CDK9 inhibition and a rapid decline in c-MYC, MCL-1, and cyclin D1 to robustly induce apoptosis, which is predominantly dependent on caspase-3 activation. Enitociclib also significantly impeded tumor growth in mouse CDX and PDX models. These data demonstrate that CDK9 is a promising target in MCL and may be utilized to overcome therapeutic resistance to BTKi and CART therapy in MCL. In a phase I dose-escalation trial, enitociclib was reported to be safe and effective in treating double-hit DLBCL patients [ 12 ]. Altogether, it highlights the targeting of CDK9 as a potentially effective regimen for treatment of advanced disease. Translational and mechanistic studies are ongoing to understand how targeting CDK9 can overcome therapeutic resistance in lymphoma.
Targeting CDK9 with the specific inhibitor enitociclib potently inhibited lymphoma cell growth by suppressing de novo expression of short-lived proteins and inducing apoptosis. (A) MYC-TARGETS-v1 was progressively enriched in BTKi-fast responders (-Fast), -slow responders (-Slow) and non-responders (-Resist) based on GSEA analysis of single-cell RNA-seq data from MCL patient samples. (B) MYC mRNA expression was higher in BTKi-resistant (BTKi-R) than BTKi-sensitive (BTKi-S) MCL cells (left panel), and its high expression correlated with poor patient survival (right panel). (C) Cell viability assay assessing the in vitro efficacy of enitociclib in 9 MCL cell lines (top panel) and 5 DLBCL cell lines (bottom panel). The IC50 is presented to the right of each cell line. (D) Enitociclib at the indicated concentrations inhibited cell viability and induced apoptosis in a dose-dependent manner in MCL cells after 24 hr of treatment. (E) Western blots show that enitociclib inhibited CDK9 phosphorylation and Pol II phosphorylation at Ser 2, reduced expression of c-MYC, MCL-1, and cyclin D1, and induced cleavage of PARP and caspase-3 in JeKo-R and Z138 cells. (F) Enitociclib dose-dependently suppressed CDK9 phosphorylation, Pol II phosphorylation, and expression of c-MYC and MCL-1 by 6 hr after treatment in JeKo-R cells. (G) Pretreatment of cycloheximide (CHX, 50?g/ml) for 1 hour diminished c-MYC expression, while pretreatment of MG132 (10?M) failed to rescue enitociclib (200 nM)-induced c-MYC downregulation in JeKo-R cells. (H) Enitociclib (200 nM) induced apoptosis at 24 and 48 hr even when the cells were treated with enitociclib for only the first 6 hr in JeKo-R cells. (I-J) Pan-caspase inhibitor Z-VAD-FMK (10?M) blocked enitociclib (200 nM)-induced apoptosis by cell apoptosis assay (I) and western blot (J) in JeKo-R cells. (K-L) Caspase-3-specific inhibitor Z-DEAD-FMK (20?M) rescued enitociclib (200 nM)-induced apoptosis by cell apoptosis assay (K) and western blot (L) in JeKo-R cells. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001
Enitociclib potently inhibited MCL cell growth in MCL cell line-derived xenografts (CDX) and patient-derived xenografts (PDX) models in mice. ( A-C ) Enitociclib (10 mg/kg, IV, twice a week) markedly inhibited tumor growth in JeKo-1 CDXs. Tumor volume ( A ), mouse survival ( B ), and mouse body weight ( C ) are plotted. Statistical significance is indicated in the graphs. ( D-L ) Enitociclib (10 mg/kg, IV, twice a week) effectively inhibited tumor growth in PDX models with ibrutinib resistance ( D-F , PDX-1), ibrutinib-venetoclax dual resistance ( G-I , PDX-2), or dual resistance to ibrutinib and CAR-T therapy ( J-L , PDX-3). Mouse tumors were dissected, imaged, and weighed ( D, G, and J ). Tumor size ( E, H, and K ), and mouse body weight ( F, I, and L ) are plotted
Data availability
The single-cell RNA sequencing dataset and bulk RNA sequencing dataset have been deposited in in the European Genome-Phenome Archive (EGA) database under the accession codes EGAS00001005019 and EGAS00001003418. All other data generated or analyzed during this study are included in this published article and its supplementary information files.
Abbreviations
Bruton’s tyrosine kinase
BTK inhibitor
chimeric antigen receptor T cell
cyclin-dependent kinase 9
cell line-derived xenograft
diffuse large B-cell lymphoma
BTKi fast responder
BTKi-resistant
BTKi non-responders
BTKi-sensitive
BTKi slow responder
BTKi-CAR-T dual resistant
Half-maximal inhibitory concentration
mantle cell lymphoma
patient-derived xenograft
RNA polymerase II
Maddocks K. Update on mantle cell lymphoma. Blood. 2018;132(16):1647–56.
Article CAS PubMed Google Scholar
Wang ML, Rule S, Martin P, Goy A, Auer R, Kahl BS, et al. Targeting BTK with ibrutinib in relapsed or refractory mantle-cell lymphoma. N Engl J Med. 2013;369(6):507–16.
Article CAS PubMed PubMed Central Google Scholar
Wang M, Rule S, Zinzani PL, Goy A, Casasnovas O, Smith SD, et al. Acalabrutinib in relapsed or refractory mantle cell lymphoma (ACE-LY-004): a single-arm, multicentre, phase 2 trial. Lancet. 2018;391(10121):659–67.
Song YQ, Zhou KS, Zou DH, Zhou JF, Hu JD, Yang HY, et al. Zanubrutinib in relapsed/refractory mantle cell lymphoma: long-term efficacy and safety results from a phase 2 study. Blood. 2022;139(21):3148–58.
Wang ML, Shah NN, Jurczak W, Zinzani PL, Eyre TA, Cheah CY, et al. Efficacy of pirtobrutinib in covalent BTK-inhibitor pre-treated relapsed/refractory mantle cell lymphoma: additional patients and extended follow-up from the phase 1/2 BRUIN study. Blood. 2022;140(Suppl 1):9368–72.
Article Google Scholar
Wang M, Munoz J, Goy A, Locke FL, Jacobson CA, Hill BT, et al. KTE-X19 CAR T-cell therapy in relapsed or refractory mantle-cell lymphoma. N Engl J Med. 2020;382(14):1331–42.
Blenk S, Engelmann JC, Pinkert S, Weniger M, Schultz J, Rosenwald A, et al. Explorative data analysis of MCL reveals gene expression networks implicated in survival and prognosis supported by explorative CGH analysis. BMC Cancer. 2008;8:106.
Article PubMed PubMed Central Google Scholar
Rosenwald A, Wright G, Wiestner A, Chan WC, Connors JM, Campo E, et al. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell. 2003;3(2):185–97.
Yan F, Jiang V, Jordan A, Che Y, Liu Y, Cai Q, et al. The HSP90-MYC-CDK9 network drives therapeutic resistance in mantle cell lymphoma. Exp Hematol Oncol. 2024;13(1):14.
Cidado J, Boiko S, Proia T, Ferguson D, Criscione SW, San Martin M, et al. AZD4573 is a highly selective CDK9 inhibitor that suppresses MCL-1 and induces apoptosis in hematologic cancer cells. Clin Cancer Res. 2020;26(4):922–34.
Byrne M, Frattini MG, Ottmann OG, Mantzaris I, Wermke M, Lee DJ, et al. Phase I study of the PTEFb inhibitor BAY 1251152 in patients with acute myelogenous leukemia. Blood. 2018;132(16):1647–56.
Google Scholar
Moreno V, Cordoba R, Morillo D, Diamond JR, Hamdy AM, Izumi R, et al. Safety and efficacy of VIP152, a CDK9 inhibitor, in patients with double-hit lymphoma (DHL). J Clin Oncol. 2021;39(15):7538.
Download references
Acknowledgements
We thank the patients and their families who contributed to this research study, and the Steve and Nancy Fox Cancer Research Fund for their generous support. We also thank Dr. Heng-Huan Lee for manuscript review and Dr. Paul Dolber for technical editing.
This work was supported by a generous gift from the Steve and Nancy Fox Cancer Research Fund.
Author information
Authors and affiliations.
Department of Lymphoma and Myeloma, The University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd, 77030, Houston, TX, USA
Vivian Jiang, William Lee, Tianci Zhang, Alexa Jordan, Fangfang Yan, Qingsong Cai, Joseph McIntosh, Jovanny Vargas, Yang Liu & Michael Wang
Department of Stem Cell Transplantation and Cellular Therapy, The University of Texas MD Anderson Cancer Center, Houston, TX, USA
Michael Wang
You can also search for this author in PubMed Google Scholar
Contributions
MW and VJ conceived the idea and designed the experiments; WL, VJ, TZ, AJ, QC, FY, JC, YL, and JM performed the experiments; WL, TZ, QC, FY, and MW performed data analysis; WL and VJ drafted the manuscript; VJ, WL, and MW revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Correspondence to Michael Wang .
Ethics declarations
Ethics approval and consent to participate.
The patient samples were collected from peripheral blood, bone marrow, or apheresis after obtaining informed consent and approval from the Institutional Review Board at The University of Texas MD Anderson Cancer Center. All animal experimental procedures and protocols were approved by the Institutional Animal Care and Use Committee of The University of Texas MD Anderson Cancer Center. According to this protocol, the maximal tumor size should be limited to 20 mm, which was not exceeded in this study.
Consent for publication
Not applicable.
Competing interests
MW has the following potential competing interests: Consultancy: AbbVie, Acerta Pharma, ADC Therapeutics America, Amphista Therapeutics Limited, AstraZeneca, Be Biopharma, BeiGene, BioInvent, Bristol Myers Squibb, Deciphera, DTRM Biopharma (Cayman) Limited, Genentech, InnoCare, Janssen, Kite Pharma, Leukemia & Lymphoma Society, Lilly, Merck, Miltenyi Biomedicine, Milken Institute, Oncternal, Parexel, Pepromene Bio, Pharmacyclics, VelosBio. Research: Acerta Pharma, AstraZeneca, BeiGene, BioInvent, Celgene, Genmab, Genentech, Innocare, Janssen, Juno Therapeutics, Kite Pharma, Lilly, Loxo Oncology, Molecular Templates, Oncternal, Pharmacyclics, VelosBio, Vincerx. Honoraria: AbbVie, Acerta Pharma, AstraZeneca, Bantam Pharmaceutical, BeiGene, BioInvent, Bristol Myers Squibb, CAHON, Dava Oncology, Eastern Virginia Medical School, Genmab, i3Health, ICML, IDEOlogy Health, Janssen, Kite Pharma, Leukemia & Lymphoma Society, Medscape, Meeting Minds Experts, MD Education, MJH Life Sciences, Merck, Moffit Cancer Center, NIH, Nurix, Oncology Specialty Group, OncLive, Pharmacyclics, Physicians Education Resources (PER), Practice Point Communications (PPC), Scripps, Studio ER Congressi, WebMD. No other authors have potential competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1
Supplementary material 2, supplementary material 3, supplementary material 4, supplementary material 5, supplementary material 6, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Jiang, V., Lee, W., Zhang, T. et al. The CDK9 inhibitor enitociclib overcomes resistance to BTK inhibition and CAR-T therapy in mantle cell lymphoma. Biomark Res 12 , 62 (2024). https://doi.org/10.1186/s40364-024-00589-7
Download citation
Received : 31 January 2024
Accepted : 04 April 2024
Published : 18 June 2024
DOI : https://doi.org/10.1186/s40364-024-00589-7
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Mantle cell lymphoma
- Enitociclib
- Therapeutic resistance
- BTK inhibitors
- CAR-T therapy.
Biomarker Research
ISSN: 2050-7771
- General enquiries: [email protected]

IMAGES
VIDEO
COMMENTS
Qualitative data transcription provides a good first step in arranging your data systematically and analyzing it. Transcription is vital for qualitative research because it: Puts qualitative data and information into a text-based format. Makes data easier to analyze and share. Allows researchers to become more immersed into the data they collect.
Transcription as an Act of Analysis. While transcription is often treated as part of the data collection process, it is also an act of analysis (Woods, 2020). When you manually transcribe an interview, for example, you make choices about how to turn the recording of the interview into text, and these decisions shape the analysis you conduct.
Transcription is indeed a crucial process in any qualitative research project as it is the first step in the analysis of raw data. I agree with Bailey that investigators should be very careful with handling this process.
Understanding the transcription process. The transcription process is a critical stage in qualitative research.It refers to the conversion of recorded or observed speech into written text, turning the fluid and dynamic nature of spoken communication into a tangible and analyzable form.In this section, we will delve deeper into the process of transcription and how it is approached in ...
The importance of transcription in research can be highlighted through several key points: Accuracy and reliability: Transcription ensures that researchers have an accurate and reliable record of spoken communication, which is essential for quality data analysis. Detailed analysis: Researchers can get deeper insights by carefully reviewing ...
For qualitative researchers, transcribing is an integral element to the research process. There are a variety of ways that researchers can approach transcription and the preparation of qualitative data for analysis. While many researchers transcribe interviews manually there are also a variety of resources that can be used in conjunction to ...
While there is a vast literature that considers the collection and analysis of qualitative data, there has been limited attention to audio transcription as part of this process. In this paper, I address this gap by discussing the main considerations, challenges and implications of audio transcription for qualitative research on the third sector. I present a framework for conducting audio ...
Transcription is a practice central to qualitative research, yet the literature that addresses transcription presents it as taken for granted in qualitative studies. In this article the author provides a review of three decades of literature on transcription between 1979 and 2009. The review establishes core understandings and issues that have ...
2.2. The transcription as a strategy in qualitative research. Most textbooks argue that transcription is a necessary step in the research process because it helps the researchers to 'know' their data and enables the crucial role of notation in conveying meaning during analysis (see MacLean et al., 2004).The first step is to figure out how much of the data the researcher would like to ...
Manual transcription by the lead researcher or team facilitates extensive engagement with the data, but it is time consuming for the individual(s) transcribing and for the project as a whole.
A key process in qualitative research data collection is the transcription of interviews, focus groups or fieldwork notes. Transcription is the process of converting audio, video or notes into accessible written format for qualitative data analysis. Transcribing can be time intensive, costly and laborious with decisions and methods of ...
Transcription is a practice central to qualitative research, yet the literature that addresses transcription presents it as taken for granted in qualitative studies. In this article the author provides a review of three decades of literature on transcription between 1979 and 2009. The review establishes core understandings and issues that have ...
Using qualitative data transcription tools and services is a game changer for many companies. Transcribing content in-house is exhausting, monotonous, and time-consuming, but a skilled transcription service can make the process quick and easy. Professional data transcription services save time.
Transforming spoken words into written text in qualitative research is a vital step in familiarizing and immersing oneself in the data. We share a three-step approach of how data transcription facilitated an interpretative act of analysis in a study using qualitative data collection methods on the barriers and facilitators of HIV testing and treatment in KwaZulu-Natal, South Africa.
Data transcription in qualitative research is the first and most crucial step of the research process (and often the most expensive). The best way to ensure a 99% accurate language data set for qualitative research studies is still through professional transcription services. However, automated transcription services are becoming more ...
Qualitative research transcription is a critical step in the research process where recorded interview audio is converted into text. According to a study by Oxford Academic, data transcription is the first step in analyzing data for qualitative research. After all, with it, you can better examine what was said in the interviews you conducted.
The process of transcription in qualitative research is a meticulous endeavor that demands a deep understanding of the communicative intricacies captured during data collection. It serves as more than just a methodological step; it is a critical activity that shapes the direction of data analysis. At its core, transcription is the act of translating oral communication into written form ...
Why accurate transcription matters in qualitative research. Precise transcription is essential to qualitative research because it supports the accuracy and essence of the whole research process. It is the first stage of data analysis and has a direct impact on the findings and recommendations of the study.
The choice between verbatim and non-verbatim transcription and whether to include timestamps depends on the research objectives, the nature of the qualitative data, and the intended analysis methods. Verbatim transcription is the go-to option for studies focused on communication patterns, emotional responses, and detailed content analysis.
Our ultimate guide to transcribe interviews for qualitative research. 1. Decide the important interview information. 2. Confirm what kind of transcript you need. 3. Have your tools ready.
Data transcription is a critical part of qualitative research, as it allows researchers to accurately capture the spoken data from interviews, focus groups and other audio or video recordings. However, transcription can be an arduous and time-consuming task, as it requires careful listening, interpretation and judgements about what to include ...
Transcription is an integral process in the qualitative analysis of language data and is widely employed in basic and applied research across a number of disciplines and in professional practice fields. Yet, methodological and theoretical issues associated with the transcription process have received scant attention in the research literature.
What is Transcription? Transcription is the process of converting spoken language into written text. It is applicable to various mediums such as audio and video recordings, even live interactions. There are two main types: Verbatim and non-verbatim. Verbatim transcription captures every word exactly as spoken.
Cathcart et al. 3 also performed in-depth profiling of DSB distribution by classifying the gene and the intergenic regions. By comparing RNA sequencing (RNA-seq) data from the earlier study, 5 DSBs were preferentially localized at the highly expressed gene regions sites, especially at the transcription start site. A motif analysis of the DSB sites identified 26 transcription factors, including ...
Data are mean ± s.e.m. Statistical analysis was performed using two-way analysis of variance (ANOVA)). ... ensuring that a physiological repertoire of transcription factors could interact with ...
Unlike animals, variability in transcription factors (TF) and their binding regions (TFBR) across the plants species is a major problem which most of the existing TFBR finding software fail to tackle, rendering them hardly of any use. This limitation has resulted into underdevelopment of plant regulatory research and rampant use of Arabidopsis like model species, generating misleading results.
Transcription factor OsNF-YC1 regulates grain size by coordinating the ... Chongqing Key Laboratory of Crop Molecular Improvement, Rice Research Institute, Academy of Agricultural Sciences, Southwest University, Chongqing, 400715 China. Zhibo Cui and Xiaowen Wang contributed equally to this work. ... All relevant data can be found within the ...
Qualitative research and transcription. While quantitative research seeks to explain, generalize and predict patterns through the analysis of variables, qualitative research questions are more interested in understanding and interpreting the socially constructed world around us (Bryman, 2016).This means that data are collected through documents, observation and interviews, and the latter are ...
Our single-cell RNA sequencing data revealed major transcriptomic reprogramming, with great enrichment of MYC-targets evolving as resistance to these therapies developed. Interestingly, cyclin-dependent kinase 9 (CDK9), a critical component of the positive transcription elongation factor-b complex, was among the top upregulated genes in Dual-R ...