- Sources of Data For Research: Types & Examples


Introduction
In the age of information, data has become the driving force behind decision-making and innovation. Whether in business, science, healthcare, or government, data serves as the foundation for insights and progress.
As a researcher, you need to understand the various sources of data as they are essential for conducting comprehensive and impactful studies. In this blog post, we will explore the primary data sources, their definitions, and examples to help you gather and analyze data effectively.
Primary Data Sources
Primary data sources refer to original data collected firsthand by researchers specifically for their research purposes. These sources provide fresh and relevant information tailored to the study’s objectives. Examples of primary data sources include surveys and questionnaires, direct observations, experiments, interviews, and focus groups. As a researcher, you must be familiar with primary data sources, which are original data collected firsthand specifically for your research purposes.
These sources hold significant value as they offer fresh and relevant information tailored to your study. Also, researchers use primary data to obtain accurate and specific insights into their research questions to confirm that the data is directly relevant to their study and meets their specific needs. Collecting primary data allows you as a researcher to control the data collection process, and monitor the data quality and reliability for their analyses and conclusions.
Examples of Primary Data Sources
- Surveys and questionnaires: Surveys and questionnaires are widely used data collection methods that allow you to gather information directly from respondents. Whether distributed online, through mail, or in person, surveys enable you to reach a large audience and collect quantitative data efficiently. However, it is crucial to design clear and unbiased questions to ensure the accuracy and reliability of responses.
- Observations: Direct observations involve systematically watching and recording events or behaviors as they occur. This method provides you with real-time data, offering unique insights into participants’ natural behavior and responses. It is particularly valuable in fields such as psychology, anthropology, and ecology, where understanding human or animal behavior is critical.
- Experiments: Experiments involve when you deliberately manipulate variables to study cause-and-effect relationships. When you control variables, your experiments provide rigorous and conclusive data, often used in scientific research. They are well-suited for hypothesis testing and determining causal relationships.
- Interviews and focus groups : Qualitative data collected through interviews and focus groups give you an in-depth exploration of participants’ opinions, beliefs, and experiences. These methods help you to understand complex issues and gain rich insights that quantitative data alone may not capture or provide for your study.
Read More: What is Primary Data? + [Examples & Collection Methods]
Secondary Data Sources
As a researcher, you should also be familiar with secondary data sources. Secondary data sources involve data collected by someone else for purposes other than your specific research. Therefore, secondary data complements primary data and can provide valuable context and insights to your research.
Examples of Secondary Data Sources
- Published literature: Published literature refers to academic papers, books, and reports published by researchers and scholars in various fields. These literatures serve as a rich source of secondary data. These sources contain valuable findings and analyses from previous studies, offering a foundation for new research and the ability to build upon existing knowledge. Reviewing published literature is essential for you to understand the current state of research in your area of study and identify gaps for further investigation.
- Government sources: Government agencies collect and maintain vast amounts of data on a wide range of topics. These datasets are often made available for public use and can be a valuable resource for researchers. For example, census data provides demographic information, economic indicators offer insights into the economy, and health records contribute to public health research. Government sources offer standardized and reliable data that can be used for various research purposes.
- Online databases: The internet has opened up access to a wealth of data through online databases, data repositories, and open data initiatives. These platforms host datasets on diverse subjects. This makes them easily accessible to you and other researchers worldwide. Online databases are particularly beneficial for conducting cross-disciplinary research or exploring topics beyond your immediate field of expertise.
- Market research reports: Market research companies conduct surveys and gather data to analyze market trends, consumer behavior, and industry insights. These reports provide valuable data for businesses and researchers seeking information on market dynamics and consumer preferences. Market research reports offer you a comprehensive view of industries and can inform you of how to make strategic decisions.
Read More: What is Secondary Data? + [Examples, Sources & Analysis]
Tertiary Data Sources
In addition to primary and secondary data, you should be aware of tertiary data sources, which play a critical role in aggregating and organizing existing data from various origins. Tertiary data sources focus on collecting, curating, and preserving data for easy access and analysis.
Examples of Tertiary Data Sources
- Data aggregators: Data aggregators are companies or organizations that specialize in collecting and compiling data from multiple sources into centralized databases. These sources can include government agencies, research institutions, businesses, and other data providers. These aggregators offer a convenient way for you, a researcher, to access a vast amount of data on specific topics or industries. As they consolidate data from diverse sources, they provide you and other researchers with a comprehensive view of trends, patterns, and insights.
- Data brokers: The best way to describe data brokers is that they are entities that buy and sell data, often without the direct consent or knowledge of the individuals whose data is being traded. While data brokers can offer access to large datasets, their practices raise privacy and ethical concerns. As a researcher, you should be cautious when using data obtained through data brokers to ensure compliance with ethical guidelines and data protection laws.
- Data archives: Data archives serve as repositories for historical data and research findings. These archives are essential for preserving valuable information for future reference and analysis. They often contain datasets, reports, academic papers, and other research materials. Data archives ensure that data remains accessible for replication studies, verification of previous research, and the development of longitudinal analyses.
Emerging Data Sources
As you delve into the world of data collection, it’s important to know the emerging sources that have gained prominence in recent years. These newer data sources provide valuable insights and opportunities for research across various domains. Below are some of these emerging data sources:
- Internet of Things (IoT): The Internet of Things (IoT) has changed data collection in the 21st century through the everyday connection of devices and objects to the Internet. Smart devices like sensors, wearables, and home appliances generate vast amounts of data in real-time. For example, IoT devices in healthcare can monitor patients’ health metrics, while in agriculture, they can optimize irrigation and crop management. As a researcher, you can leverage IoT data to analyze patterns, predict trends, and make data-driven decisions.
- Social media and web data: Social media platforms and websites host a wealth of information generated by users worldwide. When you analyze social media posts and online reviews, and scrap the web, they provide you with valuable insights into public opinions, consumer behavior, and trends. You can study sentiment analysis, track customer preferences, and identify emerging topics using social media data. Web scraping allows for the extraction of data from websites, enabling researchers to gather large datasets for analysis.
- Sensor data: Sensor data is becoming increasingly relevant in various fields, including environmental monitoring, urban planning, and healthcare. Sensors are capable of measuring and collecting data on environmental parameters, traffic patterns, air quality, and more. This data helps you understand environmental changes, optimize urban infrastructure, and improve public health initiatives. Sensor networks offer a continuous stream of data, that provides you with real-time and accurate information.
In conclusion, we have explored the diverse sources of data for research, such as primary data sources, secondary data sources, and tertiary data sources, which all play a crucial role in getting the accurate information needed for research. It is important that you understand the strengths and limitations of each data source.
As you embark on your research journey, explore and utilize these diverse data sources. And if you leverage a combination of primary, secondary, and tertiary data, you can make informed decisions, drive progress in your respective fields, and uncover novel insights that may not be achievable without trying out different sources.

Connect to Formplus, Get Started Now - It's Free!
- data sources
- primary data sources
- research studies
- secondary data source
- tertiary data source

You may also like:
Naive vs Non Naive Participants In Research: Meaning & Implications
Introduction In research studies, naive and non-naive participant information alludes to the degree of commonality and understanding...

Desk Research: Definition, Types, Application, Pros & Cons
If you are looking for a way to conduct a research study while optimizing your resources, desk research is a great option. Desk research...
Subgroup Analysis: What It Is + How to Conduct It
Introduction Clinical trials are an integral part of the drug development process. They aim to assess the safety and efficacy of a new...
Projective Techniques In Surveys: Definition, Types & Pros & Cons
Introduction When you’re conducting a survey, you need to find out what people think about things. But how do you get an accurate and...
Formplus - For Seamless Data Collection
Collect data the right way with a versatile data collection tool. try formplus and transform your work productivity today..
Data sources in research: A quick guide
Last updated
18 April 2023
Reviewed by
Cathy Heath
Whether researching for a school or academic project, to advance medical science, or to discover historical treasures, you must understand what a data source is and why data sources are important.
Analyze all your data in one place
Uncover hidden nuggets in all types of qualitative data when you analyze it in Dovetail
- What is a data source?
A data source is any location where you can find facts, figures, or other relevant information to support your research. You may create your own data source through experimentation, surveys , or observations, or you may choose to use data produced by other researchers. Both methods have advantages and disadvantages, depending on your research and the quality of the existing data you can find.
In the digital age, finding data sources has become much easier, though whether those sources will meet your research goals needs to be thoroughly investigated.
Finding reliable data sources, understanding how appropriate they are to your research, and then citing them is the researcher’s responsibility.
How do you identify data sources?
Data sources should be identified from their primary sources using bibliographic referencing. Those sources found in government, academic, and non-profit data repositories are often considered the most reliable in terms of quality.
How do you select data sources?
You should select data sources based on relevance, reliability, context, and perspective. Selecting poor data sources will ruin your chances of a successful research outcome. Think of the coding phrase: "Garbage in, garbage out."
Data sources versus reference sources
Reference sources are often a great place for researchers to begin their studies before they start taking a deeper dive into data sources to back up their own research. Reference sources offer a more expansive overview of a topic as they refer to conclusions from previous researchers’ studies.
In contrast, data sources provide the facts and figures that can drive research forward and uncover new insights.
- Why are data sources important?
In most research, you start with a hypothesis and seek to find data to support that hypothesis, or you start with an open mind about a conclusion and follow the data to where it leads. In either case, you need a large enough data set to draw conclusions and data that are relevant and accurate.
In any academic, medical, or historical field, your research will be subject to review by your peers, so data sources are important. Others must be able to repeat your research and come to the same conclusions or at least understand why you came to the conclusions.
- Examples of sources of data
Data sources abound in nearly every field of research. Databases from governmental (.gov), academic (.edu), and non-profit (.org) sources are considered more reliable than those from commercial enterprises due to possible bias—but some data from commercial areas is valid and useful.
If undertaking medical research, you can find databases from the National Institutes of Health, the Centers for Disease Control, the Federal Drug Administration, and more. These sources would be considered more authoritative than something you might find on WebMD or Wikipedia. However, you’ll also find solid research through university websites or medical facilities, such as the Mayo Clinic or MD Anderson Cancer Center.
Researchers doing historical work can access reliable data through similar sources, such as the National Archives (.gov), the Smithsonian Institution (.edu), and many academic sites from universities and colleges.
They might also find information through presidential libraries, national and local historical societies, and media and business databases.
If economic research is your thing, the Federal Reserve, the Department of Labor, and academic sites provide large research databases. Or, if you're into politics, you can source data from county voting records, state election departments, and even polling organizations such as Gallup or Harris.
How many sources of data are there?
Thousands of data sources exist, but the key is finding relevant and factual data sources to meet your research goals.
- Types of data sources
Data sources can be split into two categories: primary and secondary. Both are valid resources, depending on the type of research you’re conducting.
Primary data sources
When a researcher or research team develops their data from experimentation, surveys , or observation, these are classified as primary data sources. This research generates its own new data sets to support specific research.
Direct quantitative measurements
Experiments by researchers generate quantitative measurements in a lab, nature, or other controlled environments to test specific outcomes. For example, if agricultural researchers want to see how a certain crop grows under various conditions, they create an environment that mimics those conditions and quantifies the outcomes.
In such a case, the researchers must report how they created the conditions and measured the outcome.
In social sciences, researchers craft surveys or questionnaires to judge how people would respond to a situation. These researchers would be responsible for sampling a representative population and preparing questions without bias.
Observations
Observational researchers might count the number of species in a given environment or look at how environmental changes might affect a given species. Social researchers also use observational techniques to judge how people react to various situations.
The advantages of primary data sources
Primary data offers the advantage of giving researchers control over their entire environment and the flexibility to adapt their experiments when necessary. They can also provide answers to a new or novel situation that has never existed.
For example, when testing a newly invented medicine, researchers must experiment with animals or humans to discover its effectiveness and safety outside the test tube. They have no specific historical data to fall back on, so primary research , like a clinical trial, starts to build knowledge about that particular medicine.
Secondary data sources
Secondary data sources are those produced from previous research. In our digital world, secondary data is readily available through online databases maintained by hundreds of organizations. Using secondary data sources is less costly than primary data research and offers the advantage of speed since researchers don't need to wait for the data to play out—it's readily available.
Researchers, however, must judge whether the data applies to their particular research question , as they must account for potential bias, location, demographics, etc. The data also must be well-sourced and reliable for the researcher to make those judgments.
Databases and data repositories
Any number of agencies, organizations, or commercial enterprises maintain databases and data repositories. These generally are secure environments, with some behind paywalls, and are maintained and updated as new data becomes available.
Using publicly available data sources
Publicly available data sources abound on the Internet, allowing researchers to access information quickly. Many of these data repositories are maintained by government agencies or non-profit groups created to represent certain fields of study, such as biomedical research or physics.
It’s the role of the researcher to observe the terms of use for the data and check the methodology employed in the research to ensure it matches their research criteria.
- Understanding the context around the data
Any researcher must understand the context behind a data source before they adopt it into their research. The date of when the research was conducted, the population surveyed, and the location are key pieces of information a researcher must consider. For example, biomedical research that is ten years old might contain little useful information because the field has changed so dramatically.
But the researcher must also understand who conducted the primary research, what their motives were, and how they took their measurements. All this context would affect how the researcher can apply the data to current research.
- Perspectives on data sources to bear in mind
Researchers should craft "inclusion criteria" for what data sources will be acceptable. These rules will ensure they understand the perspective of any data sources used for the research.
Sources with a more extensive data set will be more reliable and fit better into new research.
Should you be using a customer insights hub?
Do you want to discover previous research faster?
Do you share your research findings with others?
Do you analyze research data?
Start for free today, add your research, and get to key insights faster
Editor’s picks
Last updated: 18 April 2023
Last updated: 27 February 2023
Last updated: 6 February 2023
Last updated: 15 January 2024
Last updated: 6 October 2023
Last updated: 5 February 2023
Last updated: 16 April 2023
Last updated: 7 March 2023
Last updated: 9 March 2023
Last updated: 12 December 2023
Last updated: 11 March 2024
Last updated: 13 May 2024
Latest articles
Related topics, .css-je19u9{-webkit-align-items:flex-end;-webkit-box-align:flex-end;-ms-flex-align:flex-end;align-items:flex-end;display:-webkit-box;display:-webkit-flex;display:-ms-flexbox;display:flex;-webkit-flex-direction:row;-ms-flex-direction:row;flex-direction:row;-webkit-box-flex-wrap:wrap;-webkit-flex-wrap:wrap;-ms-flex-wrap:wrap;flex-wrap:wrap;-webkit-box-pack:center;-ms-flex-pack:center;-webkit-justify-content:center;justify-content:center;row-gap:0;text-align:center;max-width:671px;}@media (max-width: 1079px){.css-je19u9{max-width:400px;}.css-je19u9>span{white-space:pre;}}@media (max-width: 799px){.css-je19u9{max-width:400px;}.css-je19u9>span{white-space:pre;}} decide what to .css-1kiodld{max-height:56px;display:-webkit-box;display:-webkit-flex;display:-ms-flexbox;display:flex;-webkit-align-items:center;-webkit-box-align:center;-ms-flex-align:center;align-items:center;}@media (max-width: 1079px){.css-1kiodld{display:none;}} build next, decide what to build next.

Users report unexpectedly high data usage, especially during streaming sessions.

Users find it hard to navigate from the home page to relevant playlists in the app.

It would be great to have a sleep timer feature, especially for bedtime listening.

I need better filters to find the songs or artists I’m looking for.
Log in or sign up
Get started for free
- Privacy Policy

Home » Primary Data – Types, Methods and Examples
Primary Data – Types, Methods and Examples
Table of Contents

Primary Data
Definition:
Primary Data refers to data that is collected firsthand by a researcher or a team of researchers for a specific research project or purpose. It is original information that has not been previously published or analyzed, and it is gathered directly from the source or through the use of data collection methods such as surveys, interviews, observations, and experiments.
Types of Primary Data
Types of Primary Data are as follows:
Surveys are one of the most common types of primary data collection methods. They involve asking a set of standardized questions to a sample of individuals or organizations, usually through a questionnaire or an online form.
Interviews involve asking open-ended or structured questions to a sample of individuals or groups in person, over the phone, or through video conferencing. They can be conducted in a one-on-one setting or in a focus group.
Observations
Observations involve systematically recording the behavior or activities of individuals or groups in a natural or controlled setting. This type of data collection is often used in fields such as anthropology, sociology, and psychology.
Experiments
Experiments involve manipulating one or more variables and observing the effects on an outcome of interest. They are commonly used in scientific research to establish cause-and-effect relationships.
Case studies
Case studies involve in-depth analysis of a particular individual, group, or organization. They typically involve collecting a variety of data, including interviews, observations, and documents.
Action research
Action research involves collecting data to improve a specific practice or process within an organization or community. It often involves collaboration between researchers and practitioners.
Formats of Primary Data
Some common formats for primary data collection include:
- Textual data : This includes written responses to surveys or interviews, as well as written notes from observations.
- Numeric data: Numeric data includes data collected through structured surveys or experiments, such as ratings, rankings, or test scores.
- Audio data : Audio data includes recordings of interviews, focus groups, or other discussions.
- Visual data: Visual data includes photographs or videos of events, behaviors, or phenomena being studied.
- Sensor data: Sensor data includes data collected through electronic sensors, such as temperature readings, GPS data, or motion data.
- Biological data : Biological data includes data collected through biological samples, such as blood, urine, or tissue samples.
Primary Data Analysis Methods
There are several methods that can be used to analyze primary data collected from research, including:
- Descriptive statistics: Descriptive statistics involve summarizing and describing the characteristics of the data collected, such as mean, median, mode, and standard deviation.
- Inferential statistics: Inferential statistics involve making inferences about a population based on a sample of data. This can include techniques such as hypothesis testing and confidence intervals.
- Qualitative analysis: Qualitative analysis involves analyzing non-numerical data, such as textual data from interviews or observations, to identify themes, patterns, or trends.
- Content analysis: Content analysis involves analyzing textual data to identify and categorize specific words or phrases, allowing researchers to identify themes or patterns in the data.
- Coding : Coding involves categorizing data into specific categories or themes, allowing researchers to identify patterns and relationships in the data.
- Data visualization : Data visualization involves creating graphs, charts, and other visual representations of data to help researchers identify patterns and relationships in the data.
Primary Data Gathering Guide
Here are some general steps to guide you in gathering primary data:
- Define your research question or problem: Clearly define the purpose of your research and the specific questions you want to answer.
- Determine the data collection method : Decide which primary data collection method(s) will be most appropriate to answer your research question or problem.
- Develop a data collection instrument : If you are using surveys or interviews, create a structured questionnaire or interview guide to ensure that you ask the same questions of all participants.
- Identify your target population : Identify the group of individuals or organizations that will provide the data you need to answer your research question or problem.
- Recruit participants: Use various methods to recruit participants, such as email, social media, or advertising.
- Collect the data : Conduct your survey, interview, observation, or experiment, ensuring that you follow your data collection instrument.
- Verify the data : Check the data for completeness, accuracy, and consistency. Resolve any missing data or errors.
- Analyze the data: Use appropriate statistical or qualitative analysis techniques to interpret the data.
- Draw conclusions: Use the results of your analysis to answer your research question or problem.
- Communicate your findings : Share your results through a written report, presentation, or publication.
Examples of Primary Data
Some real-time examples of primary data are:
- Customer surveys: When a company collects data through surveys or questionnaires, they are gathering primary data. For example, a restaurant might ask customers to rate their dining experience.
- Market research : Companies may conduct primary research to understand consumer trends or market demand. For instance, a company might conduct interviews or focus groups to gather information about consumer preferences.
- Scientific experiments: Scientists may gather primary data through experiments, such as observing the behavior of animals or testing new drugs on human subjects.
- Traffic counts: Traffic engineers might collect primary data by monitoring the flow of cars on a particular road to determine how to improve traffic flow.
- Consumer behavior : Companies may use primary data to track consumer behavior, such as how customers use a product or interact with a website.
- Social media analytics : Companies can collect primary data by analyzing social media metrics such as likes, comments, and shares to understand how their customers are engaging with their brand.
Applications of Primary Data
Primary data is useful in a wide range of applications, including research, business, and government. Here are some specific applications of primary data:
- Research : Primary data is essential for conducting scientific research, such as in fields like psychology, sociology, and biology. Researchers collect primary data through experiments, surveys, and observations.
- Marketing : Companies use primary data to understand customer needs and preferences, track consumer behavior, and develop marketing strategies. This data is typically collected through surveys, focus groups, and other market research methods.
- Business planning : Primary data can inform business decisions such as product development, pricing strategies, and expansion plans. For example, a company may gather primary data on the buying habits of its customers to decide what products to offer and how to price them.
- Public policy: Primary data is used by government agencies to develop and evaluate public policies. For example, a city government might use primary data on traffic patterns to decide where to build new roads or improve public transportation.
- Education : Primary data is used in education to evaluate student performance, identify areas of need, and develop curriculum. Teachers may gather primary data through assessments, observations, and surveys to improve their teaching methods and help students succeed.
- Healthcare : Primary data is used by healthcare professionals to diagnose and treat illnesses, track patient outcomes, and develop new treatments. Doctors and researchers collect primary data through medical tests, clinical trials, and patient surveys.
- Environmental management: Primary data is used to monitor and manage natural resources and the environment. For example, scientists and environmental managers collect primary data on water quality, air quality, and biodiversity to develop policies and programs aimed at protecting the environment.
- Product testing: Companies use primary data to test new products before they are released to the market. This data is collected through surveys, focus groups, and product testing sessions to evaluate the effectiveness and appeal of the product.
- Crime prevention : Primary data is used by law enforcement agencies to identify crime hotspots, track criminal activity, and develop crime prevention strategies. Police departments may collect primary data through crime reports, surveys, and community meetings to better understand the needs and concerns of the community.
- Disaster response: Primary data is used by emergency responders and disaster management agencies to assess the impact of disasters and develop response plans. This data is collected through surveys, interviews, and observations to identify the needs of affected populations and allocate resources accordingly.
Purpose of Primary Data
The purpose of primary data is to gather information directly from the source, without relying on secondary sources or pre-existing data. This data is collected through research methods such as surveys, interviews, experiments, and observations. Primary data is valuable because it is tailored to the specific research question or problem at hand and is collected with a specific purpose in mind. Some of the main purposes of primary data include:
- To answer research questions: Researchers use primary data to answer specific research questions, such as understanding consumer preferences, evaluating the effectiveness of a program, or testing a hypothesis.
- To gather original information : Primary data provides new and original information that is not available from other sources. This data can be used to make informed decisions, develop new products, or design new programs.
- To tailor research methods: Primary data collection methods can be customized to fit the research question or problem. This allows researchers to gather the most relevant and accurate information possible.
- To control the quality of data: Researchers have greater control over the quality of primary data, as they can design and implement the data collection methods themselves. This reduces the risk of errors or biases that may be present in secondary data sources.
- To address specific populations : Primary data can be collected from specific populations, such as customers, patients, or students. This allows researchers to gather data that is directly relevant to their research question or problem.
When to use Primary Data
Primary data should be used when the specific information required for a research question or problem cannot be obtained from existing data sources. Here are some situations where primary data would be appropriate to use:
- When no secondary data is available: Primary data should be collected when there is no existing data available that addresses the research question or problem.
- When the available secondary data is not relevant: Existing secondary data may not be specific or relevant enough to address the research question or problem at hand.
- When the research requires specific information : Primary data collection allows researchers to gather information that is tailored to their specific research question or problem.
- When the research requires a specific population: Primary data can be collected from specific populations, such as customers, patients, or employees, to provide more targeted and relevant information.
- When the research requires control over the data collection process: Primary data allows researchers to have greater control over the data collection process, which can ensure the data is of high quality and relevant to the research question or problem.
- When the research requires current or up-to-date information: Primary data collection can provide more current and up-to-date information than existing secondary data sources.
Characteristics of Primary Data
Primary data has several characteristics that make it unique and valuable for research purposes. These characteristics include:
- Originality : Primary data is collected for a specific research question or problem and is not previously published or available in any other source.
- Relevance : Primary data is collected to directly address the research question or problem at hand and is therefore highly relevant to the research.
- Accuracy : Primary data collection methods can be designed to ensure the data is accurate and reliable, reducing the risk of errors or biases.
- Timeliness: Primary data is collected in real-time or near real-time, providing current and up-to-date information for the research.
- Specificity : Primary data can be collected from specific populations, such as customers, patients, or employees, providing targeted and relevant information.
- Control : Researchers have greater control over the data collection process, allowing them to ensure the data is collected in a way that is most relevant to the research question or problem.
- Cost : Primary data collection can be more expensive than using existing secondary data sources, as it requires resources such as personnel, equipment, and materials.
Advantages of Primary Data
There are several advantages of using primary data in research. These include:
- Specificity : Primary data collection can be tailored to the specific research question or problem, allowing researchers to gather the most relevant and targeted information possible.
- Control : Researchers have greater control over the data collection process, which can ensure the data is of high quality and relevant to the research question or problem.
- Timeliness : Primary data is collected in real-time or near real-time, providing current and up-to-date information for the research.
- Flexibility : Primary data collection methods can be adjusted or modified during the research process to ensure the most relevant and useful data is collected.
- Greater depth : Primary data collection methods, such as interviews or focus groups, can provide more in-depth and detailed information than existing secondary data sources.
- Potential for new insights : Primary data collection can provide new and unexpected insights into a research question or problem, which may not have been possible using existing secondary data sources.
Limitations of Primary Data
While primary data has several advantages, it also has some limitations that researchers need to be aware of. These limitations include:
- Time-consuming: Primary data collection can be time-consuming, especially if the research requires collecting data from a large sample or a specific population.
- Limited generalizability: Primary data is collected from a specific population, and therefore its generalizability to other populations may be limited.
- Potential bias: Primary data collection methods can be subject to biases, such as social desirability bias or interviewer bias, which can affect the accuracy and reliability of the data.
- Potential for errors: Primary data collection methods can be prone to errors, such as data entry errors or measurement errors, which can affect the accuracy and reliability of the data.
- Ethical concerns: Primary data collection methods, such as interviews or surveys, may raise ethical concerns related to confidentiality, privacy, and informed consent.
About the author
Muhammad Hassan
Researcher, Academic Writer, Web developer
You may also like

Secondary Data – Types, Methods and Examples

Research Data – Types Methods and Examples

Quantitative Data – Types, Methods and Examples

Information in Research – Types and Examples

Qualitative Data – Types, Methods and Examples
Methodology and Sources of Data
- First Online: 23 July 2022
Cite this chapter

- Mohammed Moniruzzaman Khan 2
79 Accesses
Methodology is one of the approaches in which the research process is made transparent; it can be used as a strategy or a plan of action. Methodology provides grounding logic and assumptions before the design is finalised (Silverman as cited in Phellas, 2006). It helps understand why this study has been undertaken and how the research problem has been defined, what type of data has been collected, what particular method has been adopted and why a particular technique of analysing data has been used and a host of similar questions.
This is a preview of subscription content, log in via an institution to check access.
Access this chapter
- Available as PDF
- Read on any device
- Instant download
- Own it forever
- Available as EPUB and PDF
- Compact, lightweight edition
- Dispatched in 3 to 5 business days
- Free shipping worldwide - see info
- Durable hardcover edition
Tax calculation will be finalised at checkout
Purchases are for personal use only
Institutional subscriptions
A village has been established by the government on abandoned land after Aila , with a particular structure of house and a tubewell, sanitation facilities, water reservoir and community pond for people who lost their houses in Aila. The local people called it a model village.
Author information
Authors and affiliations.
Department of Sociology, Jagannath University, Dhaka, Bangladesh
Mohammed Moniruzzaman Khan
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Mohammed Moniruzzaman Khan .
Rights and permissions
Reprints and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Khan, M.M. (2022). Methodology and Sources of Data. In: Disaster and Gender in Coastal Bangladesh. Springer, Singapore. https://doi.org/10.1007/978-981-19-3284-7_2
Download citation
DOI : https://doi.org/10.1007/978-981-19-3284-7_2
Published : 23 July 2022
Publisher Name : Springer, Singapore
Print ISBN : 978-981-19-3283-0
Online ISBN : 978-981-19-3284-7
eBook Packages : Literature, Cultural and Media Studies Literature, Cultural and Media Studies (R0)
Share this chapter
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Publish with us
Policies and ethics
- Find a journal
- Track your research
News alert: UC Berkeley has announced its next university librarian
Secondary menu
- Log in to your Library account
- Hours and Maps
- Connect from Off Campus
- UC Berkeley Home
Search form
Research methods--quantitative, qualitative, and more: overview.
- Quantitative Research
- Qualitative Research
- Data Science Methods (Machine Learning, AI, Big Data)
- Text Mining and Computational Text Analysis
- Evidence Synthesis/Systematic Reviews
- Get Data, Get Help!
About Research Methods
This guide provides an overview of research methods, how to choose and use them, and supports and resources at UC Berkeley.
As Patten and Newhart note in the book Understanding Research Methods , "Research methods are the building blocks of the scientific enterprise. They are the "how" for building systematic knowledge. The accumulation of knowledge through research is by its nature a collective endeavor. Each well-designed study provides evidence that may support, amend, refute, or deepen the understanding of existing knowledge...Decisions are important throughout the practice of research and are designed to help researchers collect evidence that includes the full spectrum of the phenomenon under study, to maintain logical rules, and to mitigate or account for possible sources of bias. In many ways, learning research methods is learning how to see and make these decisions."
The choice of methods varies by discipline, by the kind of phenomenon being studied and the data being used to study it, by the technology available, and more. This guide is an introduction, but if you don't see what you need here, always contact your subject librarian, and/or take a look to see if there's a library research guide that will answer your question.
Suggestions for changes and additions to this guide are welcome!
START HERE: SAGE Research Methods
Without question, the most comprehensive resource available from the library is SAGE Research Methods. HERE IS THE ONLINE GUIDE to this one-stop shopping collection, and some helpful links are below:
- SAGE Research Methods
- Little Green Books (Quantitative Methods)
- Little Blue Books (Qualitative Methods)
- Dictionaries and Encyclopedias
- Case studies of real research projects
- Sample datasets for hands-on practice
- Streaming video--see methods come to life
- Methodspace- -a community for researchers
- SAGE Research Methods Course Mapping
Library Data Services at UC Berkeley
Library Data Services Program and Digital Scholarship Services
The LDSP offers a variety of services and tools ! From this link, check out pages for each of the following topics: discovering data, managing data, collecting data, GIS data, text data mining, publishing data, digital scholarship, open science, and the Research Data Management Program.
Be sure also to check out the visual guide to where to seek assistance on campus with any research question you may have!
Library GIS Services
Other Data Services at Berkeley
D-Lab Supports Berkeley faculty, staff, and graduate students with research in data intensive social science, including a wide range of training and workshop offerings Dryad Dryad is a simple self-service tool for researchers to use in publishing their datasets. It provides tools for the effective publication of and access to research data. Geospatial Innovation Facility (GIF) Provides leadership and training across a broad array of integrated mapping technologies on campu Research Data Management A UC Berkeley guide and consulting service for research data management issues
General Research Methods Resources
Here are some general resources for assistance:
- Assistance from ICPSR (must create an account to access): Getting Help with Data , and Resources for Students
- Wiley Stats Ref for background information on statistics topics
- Survey Documentation and Analysis (SDA) . Program for easy web-based analysis of survey data.
Consultants
- D-Lab/Data Science Discovery Consultants Request help with your research project from peer consultants.
- Research data (RDM) consulting Meet with RDM consultants before designing the data security, storage, and sharing aspects of your qualitative project.
- Statistics Department Consulting Services A service in which advanced graduate students, under faculty supervision, are available to consult during specified hours in the Fall and Spring semesters.
Related Resourcex
- IRB / CPHS Qualitative research projects with human subjects often require that you go through an ethics review.
- OURS (Office of Undergraduate Research and Scholarships) OURS supports undergraduates who want to embark on research projects and assistantships. In particular, check out their "Getting Started in Research" workshops
- Sponsored Projects Sponsored projects works with researchers applying for major external grants.
- Next: Quantitative Research >>
- Last Updated: Apr 25, 2024 11:09 AM
- URL: https://guides.lib.berkeley.edu/researchmethods
- USC Libraries
- Research Guides
Organizing Your Social Sciences Research Paper
- 6. The Methodology
- Purpose of Guide
- Design Flaws to Avoid
- Independent and Dependent Variables
- Glossary of Research Terms
- Reading Research Effectively
- Narrowing a Topic Idea
- Broadening a Topic Idea
- Extending the Timeliness of a Topic Idea
- Academic Writing Style
- Applying Critical Thinking
- Choosing a Title
- Making an Outline
- Paragraph Development
- Research Process Video Series
- Executive Summary
- The C.A.R.S. Model
- Background Information
- The Research Problem/Question
- Theoretical Framework
- Citation Tracking
- Content Alert Services
- Evaluating Sources
- Primary Sources
- Secondary Sources
- Tiertiary Sources
- Scholarly vs. Popular Publications
- Qualitative Methods
- Quantitative Methods
- Insiderness
- Using Non-Textual Elements
- Limitations of the Study
- Common Grammar Mistakes
- Writing Concisely
- Avoiding Plagiarism
- Footnotes or Endnotes?
- Further Readings
- Generative AI and Writing
- USC Libraries Tutorials and Other Guides
- Bibliography
The methods section describes actions taken to investigate a research problem and the rationale for the application of specific procedures or techniques used to identify, select, process, and analyze information applied to understanding the problem, thereby, allowing the reader to critically evaluate a study’s overall validity and reliability. The methodology section of a research paper answers two main questions: How was the data collected or generated? And, how was it analyzed? The writing should be direct and precise and always written in the past tense.
Kallet, Richard H. "How to Write the Methods Section of a Research Paper." Respiratory Care 49 (October 2004): 1229-1232.
Importance of a Good Methodology Section
You must explain how you obtained and analyzed your results for the following reasons:
- Readers need to know how the data was obtained because the method you chose affects the results and, by extension, how you interpreted their significance in the discussion section of your paper.
- Methodology is crucial for any branch of scholarship because an unreliable method produces unreliable results and, as a consequence, undermines the value of your analysis of the findings.
- In most cases, there are a variety of different methods you can choose to investigate a research problem. The methodology section of your paper should clearly articulate the reasons why you have chosen a particular procedure or technique.
- The reader wants to know that the data was collected or generated in a way that is consistent with accepted practice in the field of study. For example, if you are using a multiple choice questionnaire, readers need to know that it offered your respondents a reasonable range of answers to choose from.
- The method must be appropriate to fulfilling the overall aims of the study. For example, you need to ensure that you have a large enough sample size to be able to generalize and make recommendations based upon the findings.
- The methodology should discuss the problems that were anticipated and the steps you took to prevent them from occurring. For any problems that do arise, you must describe the ways in which they were minimized or why these problems do not impact in any meaningful way your interpretation of the findings.
- In the social and behavioral sciences, it is important to always provide sufficient information to allow other researchers to adopt or replicate your methodology. This information is particularly important when a new method has been developed or an innovative use of an existing method is utilized.
Bem, Daryl J. Writing the Empirical Journal Article. Psychology Writing Center. University of Washington; Denscombe, Martyn. The Good Research Guide: For Small-Scale Social Research Projects . 5th edition. Buckingham, UK: Open University Press, 2014; Lunenburg, Frederick C. Writing a Successful Thesis or Dissertation: Tips and Strategies for Students in the Social and Behavioral Sciences . Thousand Oaks, CA: Corwin Press, 2008.
Structure and Writing Style
I. Groups of Research Methods
There are two main groups of research methods in the social sciences:
- The e mpirical-analytical group approaches the study of social sciences in a similar manner that researchers study the natural sciences . This type of research focuses on objective knowledge, research questions that can be answered yes or no, and operational definitions of variables to be measured. The empirical-analytical group employs deductive reasoning that uses existing theory as a foundation for formulating hypotheses that need to be tested. This approach is focused on explanation.
- The i nterpretative group of methods is focused on understanding phenomenon in a comprehensive, holistic way . Interpretive methods focus on analytically disclosing the meaning-making practices of human subjects [the why, how, or by what means people do what they do], while showing how those practices arrange so that it can be used to generate observable outcomes. Interpretive methods allow you to recognize your connection to the phenomena under investigation. However, the interpretative group requires careful examination of variables because it focuses more on subjective knowledge.
II. Content
The introduction to your methodology section should begin by restating the research problem and underlying assumptions underpinning your study. This is followed by situating the methods you used to gather, analyze, and process information within the overall “tradition” of your field of study and within the particular research design you have chosen to study the problem. If the method you choose lies outside of the tradition of your field [i.e., your review of the literature demonstrates that the method is not commonly used], provide a justification for how your choice of methods specifically addresses the research problem in ways that have not been utilized in prior studies.
The remainder of your methodology section should describe the following:
- Decisions made in selecting the data you have analyzed or, in the case of qualitative research, the subjects and research setting you have examined,
- Tools and methods used to identify and collect information, and how you identified relevant variables,
- The ways in which you processed the data and the procedures you used to analyze that data, and
- The specific research tools or strategies that you utilized to study the underlying hypothesis and research questions.
In addition, an effectively written methodology section should:
- Introduce the overall methodological approach for investigating your research problem . Is your study qualitative or quantitative or a combination of both (mixed method)? Are you going to take a special approach, such as action research, or a more neutral stance?
- Indicate how the approach fits the overall research design . Your methods for gathering data should have a clear connection to your research problem. In other words, make sure that your methods will actually address the problem. One of the most common deficiencies found in research papers is that the proposed methodology is not suitable to achieving the stated objective of your paper.
- Describe the specific methods of data collection you are going to use , such as, surveys, interviews, questionnaires, observation, archival research. If you are analyzing existing data, such as a data set or archival documents, describe how it was originally created or gathered and by whom. Also be sure to explain how older data is still relevant to investigating the current research problem.
- Explain how you intend to analyze your results . Will you use statistical analysis? Will you use specific theoretical perspectives to help you analyze a text or explain observed behaviors? Describe how you plan to obtain an accurate assessment of relationships, patterns, trends, distributions, and possible contradictions found in the data.
- Provide background and a rationale for methodologies that are unfamiliar for your readers . Very often in the social sciences, research problems and the methods for investigating them require more explanation/rationale than widely accepted rules governing the natural and physical sciences. Be clear and concise in your explanation.
- Provide a justification for subject selection and sampling procedure . For instance, if you propose to conduct interviews, how do you intend to select the sample population? If you are analyzing texts, which texts have you chosen, and why? If you are using statistics, why is this set of data being used? If other data sources exist, explain why the data you chose is most appropriate to addressing the research problem.
- Provide a justification for case study selection . A common method of analyzing research problems in the social sciences is to analyze specific cases. These can be a person, place, event, phenomenon, or other type of subject of analysis that are either examined as a singular topic of in-depth investigation or multiple topics of investigation studied for the purpose of comparing or contrasting findings. In either method, you should explain why a case or cases were chosen and how they specifically relate to the research problem.
- Describe potential limitations . Are there any practical limitations that could affect your data collection? How will you attempt to control for potential confounding variables and errors? If your methodology may lead to problems you can anticipate, state this openly and show why pursuing this methodology outweighs the risk of these problems cropping up.
NOTE: Once you have written all of the elements of the methods section, subsequent revisions should focus on how to present those elements as clearly and as logically as possibly. The description of how you prepared to study the research problem, how you gathered the data, and the protocol for analyzing the data should be organized chronologically. For clarity, when a large amount of detail must be presented, information should be presented in sub-sections according to topic. If necessary, consider using appendices for raw data.
ANOTHER NOTE: If you are conducting a qualitative analysis of a research problem , the methodology section generally requires a more elaborate description of the methods used as well as an explanation of the processes applied to gathering and analyzing of data than is generally required for studies using quantitative methods. Because you are the primary instrument for generating the data [e.g., through interviews or observations], the process for collecting that data has a significantly greater impact on producing the findings. Therefore, qualitative research requires a more detailed description of the methods used.
YET ANOTHER NOTE: If your study involves interviews, observations, or other qualitative techniques involving human subjects , you may be required to obtain approval from the university's Office for the Protection of Research Subjects before beginning your research. This is not a common procedure for most undergraduate level student research assignments. However, i f your professor states you need approval, you must include a statement in your methods section that you received official endorsement and adequate informed consent from the office and that there was a clear assessment and minimization of risks to participants and to the university. This statement informs the reader that your study was conducted in an ethical and responsible manner. In some cases, the approval notice is included as an appendix to your paper.
III. Problems to Avoid
Irrelevant Detail The methodology section of your paper should be thorough but concise. Do not provide any background information that does not directly help the reader understand why a particular method was chosen, how the data was gathered or obtained, and how the data was analyzed in relation to the research problem [note: analyzed, not interpreted! Save how you interpreted the findings for the discussion section]. With this in mind, the page length of your methods section will generally be less than any other section of your paper except the conclusion.
Unnecessary Explanation of Basic Procedures Remember that you are not writing a how-to guide about a particular method. You should make the assumption that readers possess a basic understanding of how to investigate the research problem on their own and, therefore, you do not have to go into great detail about specific methodological procedures. The focus should be on how you applied a method , not on the mechanics of doing a method. An exception to this rule is if you select an unconventional methodological approach; if this is the case, be sure to explain why this approach was chosen and how it enhances the overall process of discovery.
Problem Blindness It is almost a given that you will encounter problems when collecting or generating your data, or, gaps will exist in existing data or archival materials. Do not ignore these problems or pretend they did not occur. Often, documenting how you overcame obstacles can form an interesting part of the methodology. It demonstrates to the reader that you can provide a cogent rationale for the decisions you made to minimize the impact of any problems that arose.
Literature Review Just as the literature review section of your paper provides an overview of sources you have examined while researching a particular topic, the methodology section should cite any sources that informed your choice and application of a particular method [i.e., the choice of a survey should include any citations to the works you used to help construct the survey].
It’s More than Sources of Information! A description of a research study's method should not be confused with a description of the sources of information. Such a list of sources is useful in and of itself, especially if it is accompanied by an explanation about the selection and use of the sources. The description of the project's methodology complements a list of sources in that it sets forth the organization and interpretation of information emanating from those sources.
Azevedo, L.F. et al. "How to Write a Scientific Paper: Writing the Methods Section." Revista Portuguesa de Pneumologia 17 (2011): 232-238; Blair Lorrie. “Choosing a Methodology.” In Writing a Graduate Thesis or Dissertation , Teaching Writing Series. (Rotterdam: Sense Publishers 2016), pp. 49-72; Butin, Dan W. The Education Dissertation A Guide for Practitioner Scholars . Thousand Oaks, CA: Corwin, 2010; Carter, Susan. Structuring Your Research Thesis . New York: Palgrave Macmillan, 2012; Kallet, Richard H. “How to Write the Methods Section of a Research Paper.” Respiratory Care 49 (October 2004):1229-1232; Lunenburg, Frederick C. Writing a Successful Thesis or Dissertation: Tips and Strategies for Students in the Social and Behavioral Sciences . Thousand Oaks, CA: Corwin Press, 2008. Methods Section. The Writer’s Handbook. Writing Center. University of Wisconsin, Madison; Rudestam, Kjell Erik and Rae R. Newton. “The Method Chapter: Describing Your Research Plan.” In Surviving Your Dissertation: A Comprehensive Guide to Content and Process . (Thousand Oaks, Sage Publications, 2015), pp. 87-115; What is Interpretive Research. Institute of Public and International Affairs, University of Utah; Writing the Experimental Report: Methods, Results, and Discussion. The Writing Lab and The OWL. Purdue University; Methods and Materials. The Structure, Format, Content, and Style of a Journal-Style Scientific Paper. Department of Biology. Bates College.
Writing Tip
Statistical Designs and Tests? Do Not Fear Them!
Don't avoid using a quantitative approach to analyzing your research problem just because you fear the idea of applying statistical designs and tests. A qualitative approach, such as conducting interviews or content analysis of archival texts, can yield exciting new insights about a research problem, but it should not be undertaken simply because you have a disdain for running a simple regression. A well designed quantitative research study can often be accomplished in very clear and direct ways, whereas, a similar study of a qualitative nature usually requires considerable time to analyze large volumes of data and a tremendous burden to create new paths for analysis where previously no path associated with your research problem had existed.
To locate data and statistics, GO HERE .
Another Writing Tip
Knowing the Relationship Between Theories and Methods
There can be multiple meaning associated with the term "theories" and the term "methods" in social sciences research. A helpful way to delineate between them is to understand "theories" as representing different ways of characterizing the social world when you research it and "methods" as representing different ways of generating and analyzing data about that social world. Framed in this way, all empirical social sciences research involves theories and methods, whether they are stated explicitly or not. However, while theories and methods are often related, it is important that, as a researcher, you deliberately separate them in order to avoid your theories playing a disproportionate role in shaping what outcomes your chosen methods produce.
Introspectively engage in an ongoing dialectic between the application of theories and methods to help enable you to use the outcomes from your methods to interrogate and develop new theories, or ways of framing conceptually the research problem. This is how scholarship grows and branches out into new intellectual territory.
Reynolds, R. Larry. Ways of Knowing. Alternative Microeconomics . Part 1, Chapter 3. Boise State University; The Theory-Method Relationship. S-Cool Revision. United Kingdom.
Yet Another Writing Tip
Methods and the Methodology
Do not confuse the terms "methods" and "methodology." As Schneider notes, a method refers to the technical steps taken to do research . Descriptions of methods usually include defining and stating why you have chosen specific techniques to investigate a research problem, followed by an outline of the procedures you used to systematically select, gather, and process the data [remember to always save the interpretation of data for the discussion section of your paper].
The methodology refers to a discussion of the underlying reasoning why particular methods were used . This discussion includes describing the theoretical concepts that inform the choice of methods to be applied, placing the choice of methods within the more general nature of academic work, and reviewing its relevance to examining the research problem. The methodology section also includes a thorough review of the methods other scholars have used to study the topic.
Bryman, Alan. "Of Methods and Methodology." Qualitative Research in Organizations and Management: An International Journal 3 (2008): 159-168; Schneider, Florian. “What's in a Methodology: The Difference between Method, Methodology, and Theory…and How to Get the Balance Right?” PoliticsEastAsia.com. Chinese Department, University of Leiden, Netherlands.
- << Previous: Scholarly vs. Popular Publications
- Next: Qualitative Methods >>
- Last Updated: Jun 18, 2024 10:45 AM
- URL: https://libguides.usc.edu/writingguide

Library Guides
Dissertations 4: methodology: methods.
- Introduction & Philosophy
- Methodology
Primary & Secondary Sources, Primary & Secondary Data
When describing your research methods, you can start by stating what kind of secondary and, if applicable, primary sources you used in your research. Explain why you chose such sources, how well they served your research, and identify possible issues encountered using these sources.
Definitions
There is some confusion on the use of the terms primary and secondary sources, and primary and secondary data. The confusion is also due to disciplinary differences (Lombard 2010). Whilst you are advised to consult the research methods literature in your field, we can generalise as follows:
Secondary sources
Secondary sources normally include the literature (books and articles) with the experts' findings, analysis and discussions on a certain topic (Cottrell, 2014, p123). Secondary sources often interpret primary sources.
Primary sources
Primary sources are "first-hand" information such as raw data, statistics, interviews, surveys, law statutes and law cases. Even literary texts, pictures and films can be primary sources if they are the object of research (rather than, for example, documentaries reporting on something else, in which case they would be secondary sources). The distinction between primary and secondary sources sometimes lies on the use you make of them (Cottrell, 2014, p123).
Primary data
Primary data are data (primary sources) you directly obtained through your empirical work (Saunders, Lewis and Thornhill 2015, p316).
Secondary data
Secondary data are data (primary sources) that were originally collected by someone else (Saunders, Lewis and Thornhill 2015, p316).
Comparison between primary and secondary data
| Primary data | Secondary data |
| Data collected directly | Data collected from previously done research, existing research is summarised and collated to enhance the overall effectiveness of the research. |
| Examples: Interviews (face-to-face or telephonic), Online surveys, Focus groups and Observations | Examples: data available via the internet, non-government and government agencies, public libraries, educational institutions, commercial/business information |
| Advantages: •Data collected is first hand and accurate. •Data collected can be controlled. No dilution of data. •Research method can be customized to suit personal requirements and needs of the research. | Advantages: •Information is readily available •Less expensive and less time-consuming •Quicker to conduct |
| Disadvantages: •Can be quite extensive to conduct, requiring a lot of time and resources •Sometimes one primary research method is not enough; therefore a mixed method is require, which can be even more time consuming. | Disadvantages: •It is necessary to check the credibility of the data •May not be as up to date •Success of your research depends on the quality of research previously conducted by others. |
Use
Virtually all research will use secondary sources, at least as background information.
Often, especially at the postgraduate level, it will also use primary sources - secondary and/or primary data. The engagement with primary sources is generally appreciated, as less reliant on others' interpretations, and closer to 'facts'.
The use of primary data, as opposed to secondary data, demonstrates the researcher's effort to do empirical work and find evidence to answer her specific research question and fulfill her specific research objectives. Thus, primary data contribute to the originality of the research.
Ultimately, you should state in this section of the methodology:
What sources and data you are using and why (how are they going to help you answer the research question and/or test the hypothesis.
If using primary data, why you employed certain strategies to collect them.
What the advantages and disadvantages of your strategies to collect the data (also refer to the research in you field and research methods literature).
Quantitative, Qualitative & Mixed Methods
The methodology chapter should reference your use of quantitative research, qualitative research and/or mixed methods. The following is a description of each along with their advantages and disadvantages.
Quantitative research
Quantitative research uses numerical data (quantities) deriving, for example, from experiments, closed questions in surveys, questionnaires, structured interviews or published data sets (Cottrell, 2014, p93). It normally processes and analyses this data using quantitative analysis techniques like tables, graphs and statistics to explore, present and examine relationships and trends within the data (Saunders, Lewis and Thornhill, 2015, p496).
| Advantages | Disadvantages |
| The study can be undertaken on a broader scale, generating large amounts of data that contribute to generalisation of results | Quantitative methods can be difficult, expensive and time consuming (especially if using primary data, rather than secondary data). |
| Suitable when the phenomenon is relatively simple, and can be analysed according to identified variables. | Not everything can be easily measured. |
|
| Less suitable for complex social phenomena. |
|
| Less suitable for why type questions. |
Qualitative research
Qualitative research is generally undertaken to study human behaviour and psyche. It uses methods like in-depth case studies, open-ended survey questions, unstructured interviews, focus groups, or unstructured observations (Cottrell, 2014, p93). The nature of the data is subjective, and also the analysis of the researcher involves a degree of subjective interpretation. Subjectivity can be controlled for in the research design, or has to be acknowledged as a feature of the research. Subject-specific books on (qualitative) research methods offer guidance on such research designs.
| Advantages | Disadvantages |
| Qualitative methods are good for in-depth analysis of individual people, businesses, organisations, events. | The findings can be accurate about the particular case, but not generally applicable. |
| Sample sizes don’t need to be large, so the studies can be cheaper and simpler. | More prone to subjectivity. |
Mixed methods
Mixed-method approaches combine both qualitative and quantitative methods, and therefore combine the strengths of both types of research. Mixed methods have gained popularity in recent years.
When undertaking mixed-methods research you can collect the qualitative and quantitative data either concurrently or sequentially. If sequentially, you can for example, start with a few semi-structured interviews, providing qualitative insights, and then design a questionnaire to obtain quantitative evidence that your qualitative findings can also apply to a wider population (Specht, 2019, p138).
Ultimately, your methodology chapter should state:
Whether you used quantitative research, qualitative research or mixed methods.
Why you chose such methods (and refer to research method sources).
Why you rejected other methods.
How well the method served your research.
The problems or limitations you encountered.
Doug Specht, Senior Lecturer at the Westminster School of Media and Communication, explains mixed methods research in the following video:
LinkedIn Learning Video on Academic Research Foundations: Quantitative
The video covers the characteristics of quantitative research, and explains how to approach different parts of the research process, such as creating a solid research question and developing a literature review. He goes over the elements of a study, explains how to collect and analyze data, and shows how to present your data in written and numeric form.

Link to quantitative research video
Some Types of Methods
There are several methods you can use to get primary data. To reiterate, the choice of the methods should depend on your research question/hypothesis.
Whatever methods you will use, you will need to consider:
why did you choose one technique over another? What were the advantages and disadvantages of the technique you chose?
what was the size of your sample? Who made up your sample? How did you select your sample population? Why did you choose that particular sampling strategy?)
ethical considerations (see also tab...)
safety considerations
validity
feasibility
recording
procedure of the research (see box procedural method...).
Check Stella Cottrell's book Dissertations and Project Reports: A Step by Step Guide for some succinct yet comprehensive information on most methods (the following account draws mostly on her work). Check a research methods book in your discipline for more specific guidance.
Experiments
Experiments are useful to investigate cause and effect, when the variables can be tightly controlled. They can test a theory or hypothesis in controlled conditions. Experiments do not prove or disprove an hypothesis, instead they support or not support an hypothesis. When using the empirical and inductive method it is not possible to achieve conclusive results. The results may only be valid until falsified by other experiments and observations.
For more information on Scientific Method, click here .
Observations
Observational methods are useful for in-depth analyses of behaviours in people, animals, organisations, events or phenomena. They can test a theory or products in real life or simulated settings. They generally a qualitative research method.
Questionnaires and surveys
Questionnaires and surveys are useful to gain opinions, attitudes, preferences, understandings on certain matters. They can provide quantitative data that can be collated systematically; qualitative data, if they include opportunities for open-ended responses; or both qualitative and quantitative elements.
Interviews
Interviews are useful to gain rich, qualitative information about individuals' experiences, attitudes or perspectives. With interviews you can follow up immediately on responses for clarification or further details. There are three main types of interviews: structured (following a strict pattern of questions, which expect short answers), semi-structured (following a list of questions, with the opportunity to follow up the answers with improvised questions), and unstructured (following a short list of broad questions, where the respondent can lead more the conversation) (Specht, 2019, p142).
This short video on qualitative interviews discusses best practices and covers qualitative interview design, preparation and data collection methods.
Focus groups
In this case, a group of people (normally, 4-12) is gathered for an interview where the interviewer asks questions to such group of participants. Group interactions and discussions can be highly productive, but the researcher has to beware of the group effect, whereby certain participants and views dominate the interview (Saunders, Lewis and Thornhill 2015, p419). The researcher can try to minimise this by encouraging involvement of all participants and promoting a multiplicity of views.
This video focuses on strategies for conducting research using focus groups.
Check out the guidance on online focus groups by Aliaksandr Herasimenka, which is attached at the bottom of this text box.
Case study
Case studies are often a convenient way to narrow the focus of your research by studying how a theory or literature fares with regard to a specific person, group, organisation, event or other type of entity or phenomenon you identify. Case studies can be researched using other methods, including those described in this section. Case studies give in-depth insights on the particular reality that has been examined, but may not be representative of what happens in general, they may not be generalisable, and may not be relevant to other contexts. These limitations have to be acknowledged by the researcher.
Content analysis
Content analysis consists in the study of words or images within a text. In its broad definition, texts include books, articles, essays, historical documents, speeches, conversations, advertising, interviews, social media posts, films, theatre, paintings or other visuals. Content analysis can be quantitative (e.g. word frequency) or qualitative (e.g. analysing intention and implications of the communication). It can detect propaganda, identify intentions of writers, and can see differences in types of communication (Specht, 2019, p146). Check this page on collecting, cleaning and visualising Twitter data.
Extra links and resources:
Research Methods
A clear and comprehensive overview of research methods by Emerald Publishing. It includes: crowdsourcing as a research tool; mixed methods research; case study; discourse analysis; ground theory; repertory grid; ethnographic method and participant observation; interviews; focus group; action research; analysis of qualitative data; survey design; questionnaires; statistics; experiments; empirical research; literature review; secondary data and archival materials; data collection.
Doing your dissertation during the COVID-19 pandemic
Resources providing guidance on doing dissertation research during the pandemic: Online research methods; Secondary data sources; Webinars, conferences and podcasts;
- Virtual Focus Groups Guidance on managing virtual focus groups
5 Minute Methods Videos
The following are a series of useful videos that introduce research methods in five minutes. These resources have been produced by lecturers and students with the University of Westminster's School of Media and Communication.

Case Study Research
Research Ethics
Quantitative Content Analysis
Sequential Analysis
Qualitative Content Analysis
Thematic Analysis
Social Media Research
Mixed Method Research
Procedural Method
In this part, provide an accurate, detailed account of the methods and procedures that were used in the study or the experiment (if applicable!).
Include specifics about participants, sample, materials, design and methods.
If the research involves human subjects, then include a detailed description of who and how many participated along with how the participants were selected.
Describe all materials used for the study, including equipment, written materials and testing instruments.
Identify the study's design and any variables or controls employed.
Write out the steps in the order that they were completed.
Indicate what participants were asked to do, how measurements were taken and any calculations made to raw data collected.
Specify statistical techniques applied to the data to reach your conclusions.
Provide evidence that you incorporated rigor into your research. This is the quality of being thorough and accurate and considers the logic behind your research design.
Highlight any drawbacks that may have limited your ability to conduct your research thoroughly.
You have to provide details to allow others to replicate the experiment and/or verify the data, to test the validity of the research.
Bibliography
Cottrell, S. (2014). Dissertations and project reports: a step by step guide. Hampshire, England: Palgrave Macmillan.
Lombard, E. (2010). Primary and secondary sources. The Journal of Academic Librarianship , 36(3), 250-253
Saunders, M.N.K., Lewis, P. and Thornhill, A. (2015). Research Methods for Business Students. New York: Pearson Education.
Specht, D. (2019). The Media And Communications Study Skills Student Guide . London: University of Westminster Press.
- << Previous: Introduction & Philosophy
- Next: Ethics >>
- Last Updated: Sep 14, 2022 12:58 PM
- URL: https://libguides.westminster.ac.uk/methodology-for-dissertations
CONNECT WITH US
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Can J Hosp Pharm
- v.68(3); May-Jun 2015

Qualitative Research: Data Collection, Analysis, and Management
Introduction.
In an earlier paper, 1 we presented an introduction to using qualitative research methods in pharmacy practice. In this article, we review some principles of the collection, analysis, and management of qualitative data to help pharmacists interested in doing research in their practice to continue their learning in this area. Qualitative research can help researchers to access the thoughts and feelings of research participants, which can enable development of an understanding of the meaning that people ascribe to their experiences. Whereas quantitative research methods can be used to determine how many people undertake particular behaviours, qualitative methods can help researchers to understand how and why such behaviours take place. Within the context of pharmacy practice research, qualitative approaches have been used to examine a diverse array of topics, including the perceptions of key stakeholders regarding prescribing by pharmacists and the postgraduation employment experiences of young pharmacists (see “Further Reading” section at the end of this article).
In the previous paper, 1 we outlined 3 commonly used methodologies: ethnography 2 , grounded theory 3 , and phenomenology. 4 Briefly, ethnography involves researchers using direct observation to study participants in their “real life” environment, sometimes over extended periods. Grounded theory and its later modified versions (e.g., Strauss and Corbin 5 ) use face-to-face interviews and interactions such as focus groups to explore a particular research phenomenon and may help in clarifying a less-well-understood problem, situation, or context. Phenomenology shares some features with grounded theory (such as an exploration of participants’ behaviour) and uses similar techniques to collect data, but it focuses on understanding how human beings experience their world. It gives researchers the opportunity to put themselves in another person’s shoes and to understand the subjective experiences of participants. 6 Some researchers use qualitative methodologies but adopt a different standpoint, and an example of this appears in the work of Thurston and others, 7 discussed later in this paper.
Qualitative work requires reflection on the part of researchers, both before and during the research process, as a way of providing context and understanding for readers. When being reflexive, researchers should not try to simply ignore or avoid their own biases (as this would likely be impossible); instead, reflexivity requires researchers to reflect upon and clearly articulate their position and subjectivities (world view, perspectives, biases), so that readers can better understand the filters through which questions were asked, data were gathered and analyzed, and findings were reported. From this perspective, bias and subjectivity are not inherently negative but they are unavoidable; as a result, it is best that they be articulated up-front in a manner that is clear and coherent for readers.
THE PARTICIPANT’S VIEWPOINT
What qualitative study seeks to convey is why people have thoughts and feelings that might affect the way they behave. Such study may occur in any number of contexts, but here, we focus on pharmacy practice and the way people behave with regard to medicines use (e.g., to understand patients’ reasons for nonadherence with medication therapy or to explore physicians’ resistance to pharmacists’ clinical suggestions). As we suggested in our earlier article, 1 an important point about qualitative research is that there is no attempt to generalize the findings to a wider population. Qualitative research is used to gain insights into people’s feelings and thoughts, which may provide the basis for a future stand-alone qualitative study or may help researchers to map out survey instruments for use in a quantitative study. It is also possible to use different types of research in the same study, an approach known as “mixed methods” research, and further reading on this topic may be found at the end of this paper.
The role of the researcher in qualitative research is to attempt to access the thoughts and feelings of study participants. This is not an easy task, as it involves asking people to talk about things that may be very personal to them. Sometimes the experiences being explored are fresh in the participant’s mind, whereas on other occasions reliving past experiences may be difficult. However the data are being collected, a primary responsibility of the researcher is to safeguard participants and their data. Mechanisms for such safeguarding must be clearly articulated to participants and must be approved by a relevant research ethics review board before the research begins. Researchers and practitioners new to qualitative research should seek advice from an experienced qualitative researcher before embarking on their project.
DATA COLLECTION
Whatever philosophical standpoint the researcher is taking and whatever the data collection method (e.g., focus group, one-to-one interviews), the process will involve the generation of large amounts of data. In addition to the variety of study methodologies available, there are also different ways of making a record of what is said and done during an interview or focus group, such as taking handwritten notes or video-recording. If the researcher is audio- or video-recording data collection, then the recordings must be transcribed verbatim before data analysis can begin. As a rough guide, it can take an experienced researcher/transcriber 8 hours to transcribe one 45-minute audio-recorded interview, a process than will generate 20–30 pages of written dialogue.
Many researchers will also maintain a folder of “field notes” to complement audio-taped interviews. Field notes allow the researcher to maintain and comment upon impressions, environmental contexts, behaviours, and nonverbal cues that may not be adequately captured through the audio-recording; they are typically handwritten in a small notebook at the same time the interview takes place. Field notes can provide important context to the interpretation of audio-taped data and can help remind the researcher of situational factors that may be important during data analysis. Such notes need not be formal, but they should be maintained and secured in a similar manner to audio tapes and transcripts, as they contain sensitive information and are relevant to the research. For more information about collecting qualitative data, please see the “Further Reading” section at the end of this paper.
DATA ANALYSIS AND MANAGEMENT
If, as suggested earlier, doing qualitative research is about putting oneself in another person’s shoes and seeing the world from that person’s perspective, the most important part of data analysis and management is to be true to the participants. It is their voices that the researcher is trying to hear, so that they can be interpreted and reported on for others to read and learn from. To illustrate this point, consider the anonymized transcript excerpt presented in Appendix 1 , which is taken from a research interview conducted by one of the authors (J.S.). We refer to this excerpt throughout the remainder of this paper to illustrate how data can be managed, analyzed, and presented.
Interpretation of Data
Interpretation of the data will depend on the theoretical standpoint taken by researchers. For example, the title of the research report by Thurston and others, 7 “Discordant indigenous and provider frames explain challenges in improving access to arthritis care: a qualitative study using constructivist grounded theory,” indicates at least 2 theoretical standpoints. The first is the culture of the indigenous population of Canada and the place of this population in society, and the second is the social constructivist theory used in the constructivist grounded theory method. With regard to the first standpoint, it can be surmised that, to have decided to conduct the research, the researchers must have felt that there was anecdotal evidence of differences in access to arthritis care for patients from indigenous and non-indigenous backgrounds. With regard to the second standpoint, it can be surmised that the researchers used social constructivist theory because it assumes that behaviour is socially constructed; in other words, people do things because of the expectations of those in their personal world or in the wider society in which they live. (Please see the “Further Reading” section for resources providing more information about social constructivist theory and reflexivity.) Thus, these 2 standpoints (and there may have been others relevant to the research of Thurston and others 7 ) will have affected the way in which these researchers interpreted the experiences of the indigenous population participants and those providing their care. Another standpoint is feminist standpoint theory which, among other things, focuses on marginalized groups in society. Such theories are helpful to researchers, as they enable us to think about things from a different perspective. Being aware of the standpoints you are taking in your own research is one of the foundations of qualitative work. Without such awareness, it is easy to slip into interpreting other people’s narratives from your own viewpoint, rather than that of the participants.
To analyze the example in Appendix 1 , we will adopt a phenomenological approach because we want to understand how the participant experienced the illness and we want to try to see the experience from that person’s perspective. It is important for the researcher to reflect upon and articulate his or her starting point for such analysis; for example, in the example, the coder could reflect upon her own experience as a female of a majority ethnocultural group who has lived within middle class and upper middle class settings. This personal history therefore forms the filter through which the data will be examined. This filter does not diminish the quality or significance of the analysis, since every researcher has his or her own filters; however, by explicitly stating and acknowledging what these filters are, the researcher makes it easer for readers to contextualize the work.
Transcribing and Checking
For the purposes of this paper it is assumed that interviews or focus groups have been audio-recorded. As mentioned above, transcribing is an arduous process, even for the most experienced transcribers, but it must be done to convert the spoken word to the written word to facilitate analysis. For anyone new to conducting qualitative research, it is beneficial to transcribe at least one interview and one focus group. It is only by doing this that researchers realize how difficult the task is, and this realization affects their expectations when asking others to transcribe. If the research project has sufficient funding, then a professional transcriber can be hired to do the work. If this is the case, then it is a good idea to sit down with the transcriber, if possible, and talk through the research and what the participants were talking about. This background knowledge for the transcriber is especially important in research in which people are using jargon or medical terms (as in pharmacy practice). Involving your transcriber in this way makes the work both easier and more rewarding, as he or she will feel part of the team. Transcription editing software is also available, but it is expensive. For example, ELAN (more formally known as EUDICO Linguistic Annotator, developed at the Technical University of Berlin) 8 is a tool that can help keep data organized by linking media and data files (particularly valuable if, for example, video-taping of interviews is complemented by transcriptions). It can also be helpful in searching complex data sets. Products such as ELAN do not actually automatically transcribe interviews or complete analyses, and they do require some time and effort to learn; nonetheless, for some research applications, it may be a valuable to consider such software tools.
All audio recordings should be transcribed verbatim, regardless of how intelligible the transcript may be when it is read back. Lines of text should be numbered. Once the transcription is complete, the researcher should read it while listening to the recording and do the following: correct any spelling or other errors; anonymize the transcript so that the participant cannot be identified from anything that is said (e.g., names, places, significant events); insert notations for pauses, laughter, looks of discomfort; insert any punctuation, such as commas and full stops (periods) (see Appendix 1 for examples of inserted punctuation), and include any other contextual information that might have affected the participant (e.g., temperature or comfort of the room).
Dealing with the transcription of a focus group is slightly more difficult, as multiple voices are involved. One way of transcribing such data is to “tag” each voice (e.g., Voice A, Voice B). In addition, the focus group will usually have 2 facilitators, whose respective roles will help in making sense of the data. While one facilitator guides participants through the topic, the other can make notes about context and group dynamics. More information about group dynamics and focus groups can be found in resources listed in the “Further Reading” section.
Reading between the Lines
During the process outlined above, the researcher can begin to get a feel for the participant’s experience of the phenomenon in question and can start to think about things that could be pursued in subsequent interviews or focus groups (if appropriate). In this way, one participant’s narrative informs the next, and the researcher can continue to interview until nothing new is being heard or, as it says in the text books, “saturation is reached”. While continuing with the processes of coding and theming (described in the next 2 sections), it is important to consider not just what the person is saying but also what they are not saying. For example, is a lengthy pause an indication that the participant is finding the subject difficult, or is the person simply deciding what to say? The aim of the whole process from data collection to presentation is to tell the participants’ stories using exemplars from their own narratives, thus grounding the research findings in the participants’ lived experiences.
Smith 9 suggested a qualitative research method known as interpretative phenomenological analysis, which has 2 basic tenets: first, that it is rooted in phenomenology, attempting to understand the meaning that individuals ascribe to their lived experiences, and second, that the researcher must attempt to interpret this meaning in the context of the research. That the researcher has some knowledge and expertise in the subject of the research means that he or she can have considerable scope in interpreting the participant’s experiences. Larkin and others 10 discussed the importance of not just providing a description of what participants say. Rather, interpretative phenomenological analysis is about getting underneath what a person is saying to try to truly understand the world from his or her perspective.
Once all of the research interviews have been transcribed and checked, it is time to begin coding. Field notes compiled during an interview can be a useful complementary source of information to facilitate this process, as the gap in time between an interview, transcribing, and coding can result in memory bias regarding nonverbal or environmental context issues that may affect interpretation of data.
Coding refers to the identification of topics, issues, similarities, and differences that are revealed through the participants’ narratives and interpreted by the researcher. This process enables the researcher to begin to understand the world from each participant’s perspective. Coding can be done by hand on a hard copy of the transcript, by making notes in the margin or by highlighting and naming sections of text. More commonly, researchers use qualitative research software (e.g., NVivo, QSR International Pty Ltd; www.qsrinternational.com/products_nvivo.aspx ) to help manage their transcriptions. It is advised that researchers undertake a formal course in the use of such software or seek supervision from a researcher experienced in these tools.
Returning to Appendix 1 and reading from lines 8–11, a code for this section might be “diagnosis of mental health condition”, but this would just be a description of what the participant is talking about at that point. If we read a little more deeply, we can ask ourselves how the participant might have come to feel that the doctor assumed he or she was aware of the diagnosis or indeed that they had only just been told the diagnosis. There are a number of pauses in the narrative that might suggest the participant is finding it difficult to recall that experience. Later in the text, the participant says “nobody asked me any questions about my life” (line 19). This could be coded simply as “health care professionals’ consultation skills”, but that would not reflect how the participant must have felt never to be asked anything about his or her personal life, about the participant as a human being. At the end of this excerpt, the participant just trails off, recalling that no-one showed any interest, which makes for very moving reading. For practitioners in pharmacy, it might also be pertinent to explore the participant’s experience of akathisia and why this was left untreated for 20 years.
One of the questions that arises about qualitative research relates to the reliability of the interpretation and representation of the participants’ narratives. There are no statistical tests that can be used to check reliability and validity as there are in quantitative research. However, work by Lincoln and Guba 11 suggests that there are other ways to “establish confidence in the ‘truth’ of the findings” (p. 218). They call this confidence “trustworthiness” and suggest that there are 4 criteria of trustworthiness: credibility (confidence in the “truth” of the findings), transferability (showing that the findings have applicability in other contexts), dependability (showing that the findings are consistent and could be repeated), and confirmability (the extent to which the findings of a study are shaped by the respondents and not researcher bias, motivation, or interest).
One way of establishing the “credibility” of the coding is to ask another researcher to code the same transcript and then to discuss any similarities and differences in the 2 resulting sets of codes. This simple act can result in revisions to the codes and can help to clarify and confirm the research findings.
Theming refers to the drawing together of codes from one or more transcripts to present the findings of qualitative research in a coherent and meaningful way. For example, there may be examples across participants’ narratives of the way in which they were treated in hospital, such as “not being listened to” or “lack of interest in personal experiences” (see Appendix 1 ). These may be drawn together as a theme running through the narratives that could be named “the patient’s experience of hospital care”. The importance of going through this process is that at its conclusion, it will be possible to present the data from the interviews using quotations from the individual transcripts to illustrate the source of the researchers’ interpretations. Thus, when the findings are organized for presentation, each theme can become the heading of a section in the report or presentation. Underneath each theme will be the codes, examples from the transcripts, and the researcher’s own interpretation of what the themes mean. Implications for real life (e.g., the treatment of people with chronic mental health problems) should also be given.
DATA SYNTHESIS
In this final section of this paper, we describe some ways of drawing together or “synthesizing” research findings to represent, as faithfully as possible, the meaning that participants ascribe to their life experiences. This synthesis is the aim of the final stage of qualitative research. For most readers, the synthesis of data presented by the researcher is of crucial significance—this is usually where “the story” of the participants can be distilled, summarized, and told in a manner that is both respectful to those participants and meaningful to readers. There are a number of ways in which researchers can synthesize and present their findings, but any conclusions drawn by the researchers must be supported by direct quotations from the participants. In this way, it is made clear to the reader that the themes under discussion have emerged from the participants’ interviews and not the mind of the researcher. The work of Latif and others 12 gives an example of how qualitative research findings might be presented.
Planning and Writing the Report
As has been suggested above, if researchers code and theme their material appropriately, they will naturally find the headings for sections of their report. Qualitative researchers tend to report “findings” rather than “results”, as the latter term typically implies that the data have come from a quantitative source. The final presentation of the research will usually be in the form of a report or a paper and so should follow accepted academic guidelines. In particular, the article should begin with an introduction, including a literature review and rationale for the research. There should be a section on the chosen methodology and a brief discussion about why qualitative methodology was most appropriate for the study question and why one particular methodology (e.g., interpretative phenomenological analysis rather than grounded theory) was selected to guide the research. The method itself should then be described, including ethics approval, choice of participants, mode of recruitment, and method of data collection (e.g., semistructured interviews or focus groups), followed by the research findings, which will be the main body of the report or paper. The findings should be written as if a story is being told; as such, it is not necessary to have a lengthy discussion section at the end. This is because much of the discussion will take place around the participants’ quotes, such that all that is needed to close the report or paper is a summary, limitations of the research, and the implications that the research has for practice. As stated earlier, it is not the intention of qualitative research to allow the findings to be generalized, and therefore this is not, in itself, a limitation.
Planning out the way that findings are to be presented is helpful. It is useful to insert the headings of the sections (the themes) and then make a note of the codes that exemplify the thoughts and feelings of your participants. It is generally advisable to put in the quotations that you want to use for each theme, using each quotation only once. After all this is done, the telling of the story can begin as you give your voice to the experiences of the participants, writing around their quotations. Do not be afraid to draw assumptions from the participants’ narratives, as this is necessary to give an in-depth account of the phenomena in question. Discuss these assumptions, drawing on your participants’ words to support you as you move from one code to another and from one theme to the next. Finally, as appropriate, it is possible to include examples from literature or policy documents that add support for your findings. As an exercise, you may wish to code and theme the sample excerpt in Appendix 1 and tell the participant’s story in your own way. Further reading about “doing” qualitative research can be found at the end of this paper.
CONCLUSIONS
Qualitative research can help researchers to access the thoughts and feelings of research participants, which can enable development of an understanding of the meaning that people ascribe to their experiences. It can be used in pharmacy practice research to explore how patients feel about their health and their treatment. Qualitative research has been used by pharmacists to explore a variety of questions and problems (see the “Further Reading” section for examples). An understanding of these issues can help pharmacists and other health care professionals to tailor health care to match the individual needs of patients and to develop a concordant relationship. Doing qualitative research is not easy and may require a complete rethink of how research is conducted, particularly for researchers who are more familiar with quantitative approaches. There are many ways of conducting qualitative research, and this paper has covered some of the practical issues regarding data collection, analysis, and management. Further reading around the subject will be essential to truly understand this method of accessing peoples’ thoughts and feelings to enable researchers to tell participants’ stories.
Appendix 1. Excerpt from a sample transcript
The participant (age late 50s) had suffered from a chronic mental health illness for 30 years. The participant had become a “revolving door patient,” someone who is frequently in and out of hospital. As the participant talked about past experiences, the researcher asked:
- What was treatment like 30 years ago?
- Umm—well it was pretty much they could do what they wanted with you because I was put into the er, the er kind of system er, I was just on
- endless section threes.
- Really…
- But what I didn’t realize until later was that if you haven’t actually posed a threat to someone or yourself they can’t really do that but I didn’t know
- that. So wh-when I first went into hospital they put me on the forensic ward ’cause they said, “We don’t think you’ll stay here we think you’ll just
- run-run away.” So they put me then onto the acute admissions ward and – er – I can remember one of the first things I recall when I got onto that
- ward was sitting down with a er a Dr XXX. He had a book this thick [gestures] and on each page it was like three questions and he went through
- all these questions and I answered all these questions. So we’re there for I don’t maybe two hours doing all that and he asked me he said “well
- when did somebody tell you then that you have schizophrenia” I said “well nobody’s told me that” so he seemed very surprised but nobody had
- actually [pause] whe-when I first went up there under police escort erm the senior kind of consultants people I’d been to where I was staying and
- ermm so er [pause] I . . . the, I can remember the very first night that I was there and given this injection in this muscle here [gestures] and just
- having dreadful side effects the next day I woke up [pause]
- . . . and I suffered that akathesia I swear to you, every minute of every day for about 20 years.
- Oh how awful.
- And that side of it just makes life impossible so the care on the wards [pause] umm I don’t know it’s kind of, it’s kind of hard to put into words
- [pause]. Because I’m not saying they were sort of like not friendly or interested but then nobody ever seemed to want to talk about your life [pause]
- nobody asked me any questions about my life. The only questions that came into was they asked me if I’d be a volunteer for these student exams
- and things and I said “yeah” so all the questions were like “oh what jobs have you done,” er about your relationships and things and er but
- nobody actually sat down and had a talk and showed some interest in you as a person you were just there basically [pause] um labelled and you
- know there was there was [pause] but umm [pause] yeah . . .
This article is the 10th in the CJHP Research Primer Series, an initiative of the CJHP Editorial Board and the CSHP Research Committee. The planned 2-year series is intended to appeal to relatively inexperienced researchers, with the goal of building research capacity among practising pharmacists. The articles, presenting simple but rigorous guidance to encourage and support novice researchers, are being solicited from authors with appropriate expertise.
Previous articles in this series:
Bond CM. The research jigsaw: how to get started. Can J Hosp Pharm . 2014;67(1):28–30.
Tully MP. Research: articulating questions, generating hypotheses, and choosing study designs. Can J Hosp Pharm . 2014;67(1):31–4.
Loewen P. Ethical issues in pharmacy practice research: an introductory guide. Can J Hosp Pharm. 2014;67(2):133–7.
Tsuyuki RT. Designing pharmacy practice research trials. Can J Hosp Pharm . 2014;67(3):226–9.
Bresee LC. An introduction to developing surveys for pharmacy practice research. Can J Hosp Pharm . 2014;67(4):286–91.
Gamble JM. An introduction to the fundamentals of cohort and case–control studies. Can J Hosp Pharm . 2014;67(5):366–72.
Austin Z, Sutton J. Qualitative research: getting started. C an J Hosp Pharm . 2014;67(6):436–40.
Houle S. An introduction to the fundamentals of randomized controlled trials in pharmacy research. Can J Hosp Pharm . 2014; 68(1):28–32.
Charrois TL. Systematic reviews: What do you need to know to get started? Can J Hosp Pharm . 2014;68(2):144–8.
Competing interests: None declared.
Further Reading
Examples of qualitative research in pharmacy practice.
- Farrell B, Pottie K, Woodend K, Yao V, Dolovich L, Kennie N, et al. Shifts in expectations: evaluating physicians’ perceptions as pharmacists integrated into family practice. J Interprof Care. 2010; 24 (1):80–9. [ PubMed ] [ Google Scholar ]
- Gregory P, Austin Z. Postgraduation employment experiences of new pharmacists in Ontario in 2012–2013. Can Pharm J. 2014; 147 (5):290–9. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Marks PZ, Jennnings B, Farrell B, Kennie-Kaulbach N, Jorgenson D, Pearson-Sharpe J, et al. “I gained a skill and a change in attitude”: a case study describing how an online continuing professional education course for pharmacists supported achievement of its transfer to practice outcomes. Can J Univ Contin Educ. 2014; 40 (2):1–18. [ Google Scholar ]
- Nair KM, Dolovich L, Brazil K, Raina P. It’s all about relationships: a qualitative study of health researchers’ perspectives on interdisciplinary research. BMC Health Serv Res. 2008; 8 :110. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Pojskic N, MacKeigan L, Boon H, Austin Z. Initial perceptions of key stakeholders in Ontario regarding independent prescriptive authority for pharmacists. Res Soc Adm Pharm. 2014; 10 (2):341–54. [ PubMed ] [ Google Scholar ]
Qualitative Research in General
- Breakwell GM, Hammond S, Fife-Schaw C. Research methods in psychology. Thousand Oaks (CA): Sage Publications; 1995. [ Google Scholar ]
- Given LM. 100 questions (and answers) about qualitative research. Thousand Oaks (CA): Sage Publications; 2015. [ Google Scholar ]
- Miles B, Huberman AM. Qualitative data analysis. Thousand Oaks (CA): Sage Publications; 2009. [ Google Scholar ]
- Patton M. Qualitative research and evaluation methods. Thousand Oaks (CA): Sage Publications; 2002. [ Google Scholar ]
- Willig C. Introducing qualitative research in psychology. Buckingham (UK): Open University Press; 2001. [ Google Scholar ]
Group Dynamics in Focus Groups
- Farnsworth J, Boon B. Analysing group dynamics within the focus group. Qual Res. 2010; 10 (5):605–24. [ Google Scholar ]
Social Constructivism
- Social constructivism. Berkeley (CA): University of California, Berkeley, Berkeley Graduate Division, Graduate Student Instruction Teaching & Resource Center; [cited 2015 June 4]. Available from: http://gsi.berkeley.edu/gsi-guide-contents/learning-theory-research/social-constructivism/ [ Google Scholar ]
Mixed Methods
- Creswell J. Research design: qualitative, quantitative, and mixed methods approaches. Thousand Oaks (CA): Sage Publications; 2009. [ Google Scholar ]
Collecting Qualitative Data
- Arksey H, Knight P. Interviewing for social scientists: an introductory resource with examples. Thousand Oaks (CA): Sage Publications; 1999. [ Google Scholar ]
- Guest G, Namey EE, Mitchel ML. Collecting qualitative data: a field manual for applied research. Thousand Oaks (CA): Sage Publications; 2013. [ Google Scholar ]
Constructivist Grounded Theory
- Charmaz K. Grounded theory: objectivist and constructivist methods. In: Denzin N, Lincoln Y, editors. Handbook of qualitative research. 2nd ed. Thousand Oaks (CA): Sage Publications; 2000. pp. 509–35. [ Google Scholar ]
Have a language expert improve your writing
Run a free plagiarism check in 10 minutes, automatically generate references for free.
- Knowledge Base
- Methodology
- Data Collection Methods | Step-by-Step Guide & Examples
Data Collection Methods | Step-by-Step Guide & Examples
Published on 4 May 2022 by Pritha Bhandari .
Data collection is a systematic process of gathering observations or measurements. Whether you are performing research for business, governmental, or academic purposes, data collection allows you to gain first-hand knowledge and original insights into your research problem .
While methods and aims may differ between fields, the overall process of data collection remains largely the same. Before you begin collecting data, you need to consider:
- The aim of the research
- The type of data that you will collect
- The methods and procedures you will use to collect, store, and process the data
To collect high-quality data that is relevant to your purposes, follow these four steps.

Table of contents
Step 1: define the aim of your research, step 2: choose your data collection method, step 3: plan your data collection procedures, step 4: collect the data, frequently asked questions about data collection.
Before you start the process of data collection, you need to identify exactly what you want to achieve. You can start by writing a problem statement : what is the practical or scientific issue that you want to address, and why does it matter?
Next, formulate one or more research questions that precisely define what you want to find out. Depending on your research questions, you might need to collect quantitative or qualitative data :
- Quantitative data is expressed in numbers and graphs and is analysed through statistical methods .
- Qualitative data is expressed in words and analysed through interpretations and categorisations.
If your aim is to test a hypothesis , measure something precisely, or gain large-scale statistical insights, collect quantitative data. If your aim is to explore ideas, understand experiences, or gain detailed insights into a specific context, collect qualitative data.
If you have several aims, you can use a mixed methods approach that collects both types of data.
- Your first aim is to assess whether there are significant differences in perceptions of managers across different departments and office locations.
- Your second aim is to gather meaningful feedback from employees to explore new ideas for how managers can improve.
Prevent plagiarism, run a free check.
Based on the data you want to collect, decide which method is best suited for your research.
- Experimental research is primarily a quantitative method.
- Interviews , focus groups , and ethnographies are qualitative methods.
- Surveys , observations, archival research, and secondary data collection can be quantitative or qualitative methods.
Carefully consider what method you will use to gather data that helps you directly answer your research questions.
| Method | When to use | How to collect data |
|---|---|---|
| Experiment | To test a causal relationship. | Manipulate variables and measure their effects on others. |
| Survey | To understand the general characteristics or opinions of a group of people. | Distribute a list of questions to a sample online, in person, or over the phone. |
| Interview/focus group | To gain an in-depth understanding of perceptions or opinions on a topic. | Verbally ask participants open-ended questions in individual interviews or focus group discussions. |
| Observation | To understand something in its natural setting. | Measure or survey a sample without trying to affect them. |
| Ethnography | To study the culture of a community or organisation first-hand. | Join and participate in a community and record your observations and reflections. |
| Archival research | To understand current or historical events, conditions, or practices. | Access manuscripts, documents, or records from libraries, depositories, or the internet. |
| Secondary data collection | To analyse data from populations that you can’t access first-hand. | Find existing datasets that have already been collected, from sources such as government agencies or research organisations. |
When you know which method(s) you are using, you need to plan exactly how you will implement them. What procedures will you follow to make accurate observations or measurements of the variables you are interested in?
For instance, if you’re conducting surveys or interviews, decide what form the questions will take; if you’re conducting an experiment, make decisions about your experimental design .
Operationalisation
Sometimes your variables can be measured directly: for example, you can collect data on the average age of employees simply by asking for dates of birth. However, often you’ll be interested in collecting data on more abstract concepts or variables that can’t be directly observed.
Operationalisation means turning abstract conceptual ideas into measurable observations. When planning how you will collect data, you need to translate the conceptual definition of what you want to study into the operational definition of what you will actually measure.
- You ask managers to rate their own leadership skills on 5-point scales assessing the ability to delegate, decisiveness, and dependability.
- You ask their direct employees to provide anonymous feedback on the managers regarding the same topics.
You may need to develop a sampling plan to obtain data systematically. This involves defining a population , the group you want to draw conclusions about, and a sample, the group you will actually collect data from.
Your sampling method will determine how you recruit participants or obtain measurements for your study. To decide on a sampling method you will need to consider factors like the required sample size, accessibility of the sample, and time frame of the data collection.
Standardising procedures
If multiple researchers are involved, write a detailed manual to standardise data collection procedures in your study.
This means laying out specific step-by-step instructions so that everyone in your research team collects data in a consistent way – for example, by conducting experiments under the same conditions and using objective criteria to record and categorise observations.
This helps ensure the reliability of your data, and you can also use it to replicate the study in the future.
Creating a data management plan
Before beginning data collection, you should also decide how you will organise and store your data.
- If you are collecting data from people, you will likely need to anonymise and safeguard the data to prevent leaks of sensitive information (e.g. names or identity numbers).
- If you are collecting data via interviews or pencil-and-paper formats, you will need to perform transcriptions or data entry in systematic ways to minimise distortion.
- You can prevent loss of data by having an organisation system that is routinely backed up.
Finally, you can implement your chosen methods to measure or observe the variables you are interested in.
The closed-ended questions ask participants to rate their manager’s leadership skills on scales from 1 to 5. The data produced is numerical and can be statistically analysed for averages and patterns.
To ensure that high-quality data is recorded in a systematic way, here are some best practices:
- Record all relevant information as and when you obtain data. For example, note down whether or how lab equipment is recalibrated during an experimental study.
- Double-check manual data entry for errors.
- If you collect quantitative data, you can assess the reliability and validity to get an indication of your data quality.
Data collection is the systematic process by which observations or measurements are gathered in research. It is used in many different contexts by academics, governments, businesses, and other organisations.
When conducting research, collecting original data has significant advantages:
- You can tailor data collection to your specific research aims (e.g., understanding the needs of your consumers or user testing your website).
- You can control and standardise the process for high reliability and validity (e.g., choosing appropriate measurements and sampling methods ).
However, there are also some drawbacks: data collection can be time-consuming, labour-intensive, and expensive. In some cases, it’s more efficient to use secondary data that has already been collected by someone else, but the data might be less reliable.
Quantitative research deals with numbers and statistics, while qualitative research deals with words and meanings.
Quantitative methods allow you to test a hypothesis by systematically collecting and analysing data, while qualitative methods allow you to explore ideas and experiences in depth.
Reliability and validity are both about how well a method measures something:
- Reliability refers to the consistency of a measure (whether the results can be reproduced under the same conditions).
- Validity refers to the accuracy of a measure (whether the results really do represent what they are supposed to measure).
If you are doing experimental research , you also have to consider the internal and external validity of your experiment.
In mixed methods research , you use both qualitative and quantitative data collection and analysis methods to answer your research question .
Operationalisation means turning abstract conceptual ideas into measurable observations.
For example, the concept of social anxiety isn’t directly observable, but it can be operationally defined in terms of self-rating scores, behavioural avoidance of crowded places, or physical anxiety symptoms in social situations.
Before collecting data , it’s important to consider how you will operationalise the variables that you want to measure.
Cite this Scribbr article
If you want to cite this source, you can copy and paste the citation or click the ‘Cite this Scribbr article’ button to automatically add the citation to our free Reference Generator.
Bhandari, P. (2022, May 04). Data Collection Methods | Step-by-Step Guide & Examples. Scribbr. Retrieved 18 June 2024, from https://www.scribbr.co.uk/research-methods/data-collection-guide/
Is this article helpful?

Pritha Bhandari
Other students also liked, qualitative vs quantitative research | examples & methods, triangulation in research | guide, types, examples, what is a conceptual framework | tips & examples.

Higher Education News , Tips for Online Students , Tips for Students
A Comprehensive Guide to Different Types of Research
Updated: June 19, 2024
Published: June 15, 2024

When embarking on a research project, selecting the right methodology can be the difference between success and failure. With various methods available, each suited to different types of research, it’s essential you make an informed choice. This blog post will provide tips on how to choose a research methodology that best fits your research goals .
We’ll start with definitions: Research is the systematic process of exploring, investigating, and discovering new information or validating existing knowledge. It involves defining questions, collecting data, analyzing results, and drawing conclusions.
Meanwhile, a research methodology is a structured plan that outlines how your research is to be conducted. A complete methodology should detail the strategies, processes, and techniques you plan to use for your data collection and analysis.

Research Methods
The first step of a research methodology is to identify a focused research topic, which is the question you seek to answer. By setting clear boundaries on the scope of your research, you can concentrate on specific aspects of a problem without being overwhelmed by information. This will produce more accurate findings.
Along with clarifying your research topic, your methodology should also address your research methods. Let’s look at the four main types of research: descriptive, correlational, experimental, and diagnostic.
Descriptive Research
Descriptive research is an approach designed to describe the characteristics of a population systematically and accurately. This method focuses on answering “what” questions by providing detailed observations about the subject. Descriptive research employs surveys, observational studies , and case studies to gather qualitative or quantitative data.
A real-world example of descriptive research is a survey investigating consumer behavior toward a competitor’s product. By analyzing the survey results, the company can gather detailed insights into how consumers perceive a competitor’s product, which can inform their marketing strategies and product development.
Correlational Research
Correlational research examines the statistical relationship between two or more variables to determine whether a relationship exists. Correlational research is particularly useful when ethical or practical constraints prevent experimental manipulation. It is often employed in fields such as psychology, education, and health sciences to provide insights into complex real-world interactions, helping to develop theories and inform further experimental research.
An example of correlational research is the study of the relationship between smoking and lung cancer. Researchers observe and collect data on individuals’ smoking habits and the incidence of lung cancer to determine if there is a correlation between the two variables. This type of research helps identify patterns and relationships, indicating whether increased smoking is associated with higher rates of lung cancer.
Experimental Research
Experimental research is a scientific approach where researchers manipulate one or more independent variables to observe their effect on a dependent variable. This method is designed to establish cause-and-effect relationships. Fields like psychology , medicine, and social sciences frequently employ experimental research to test hypotheses and theories under controlled conditions.
A real-world example of experimental research is Pavlov’s Dog experiment. In this experiment, Ivan Pavlov demonstrated classical conditioning by ringing a bell each time he fed his dogs. After repeating this process multiple times, the dogs began to salivate just by hearing the bell, even when no food was presented. This experiment helped to illustrate how certain stimuli can elicit specific responses through associative learning.
Diagnostic Research
Diagnostic research tries to accurately diagnose a problem by identifying its underlying causes. This type of research is crucial for understanding complex situations where a precise diagnosis is necessary for formulating effective solutions. It involves methods such as case studies and data analysis and often integrates both qualitative and quantitative data to provide a comprehensive view of the issue at hand.
An example of diagnostic research is studying the causes of a specific illness outbreak. During an outbreak of a respiratory virus, researchers might conduct diagnostic research to determine the factors contributing to the spread of the virus. This could involve analyzing patient data, testing environmental samples, and evaluating potential sources of infection. The goal is to identify the root causes and contributing factors to develop effective containment and prevention strategies.
Using an established research method is imperative, no matter if you are researching for marketing , technology , healthcare , engineering, or social science. A methodology lends legitimacy to your research by ensuring your data is both consistent and credible. A well-defined methodology also enhances the reliability and validity of the research findings, which is crucial for drawing accurate and meaningful conclusions.
Additionally, methodologies help researchers stay focused and on track, limiting the scope of the study to relevant questions and objectives. This not only improves the quality of the research but also ensures that the study can be replicated and verified by other researchers, further solidifying its scientific value.

How to Choose a Research Methodology
Choosing the best research methodology for your project involves several key steps to ensure that your approach aligns with your research goals and questions. Here’s a simplified guide to help you make the best choice.
Understand Your Goals
Clearly define the objectives of your research. What do you aim to discover, prove, or understand? Understanding your goals helps in selecting a methodology that aligns with your research purpose.
Consider the Nature of Your Data
Determine whether your research will involve numerical data, textual data, or both. Quantitative methods are best for numerical data, while qualitative methods are suitable for textual or thematic data.
Understand the Purpose of Each Methodology
Becoming familiar with the four types of research – descriptive, correlational, experimental, and diagnostic – will enable you to select the most appropriate method for your research. Many times, you will want to use a combination of methods to gather meaningful data.
Evaluate Resources and Constraints
Consider the resources available to you, including time, budget, and access to data. Some methodologies may require more resources or longer timeframes to implement effectively.
Review Similar Studies
Look at previous research in your field to see which methodologies were successful. This can provide insights and help you choose a proven approach.
By following these steps, you can select a research methodology that best fits your project’s requirements and ensures robust, credible results.
Completing Your Research Project
Upon completing your research, the next critical step is to analyze and interpret the data you’ve collected. This involves summarizing the key findings, identifying patterns, and determining how these results address your initial research questions. By thoroughly examining the data, you can draw meaningful conclusions that contribute to the body of knowledge in your field.
It’s essential that you present these findings clearly and concisely, using charts, graphs, and tables to enhance comprehension. Furthermore, discuss the implications of your results, any limitations encountered during the study, and how your findings align with or challenge existing theories.
Your research project should conclude with a strong statement that encapsulates the essence of your research and its broader impact. This final section should leave readers with a clear understanding of the value of your work and inspire continued exploration and discussion in the field.
Now that you know how to perform quality research , it’s time to get started! Applying the right research methodologies can make a significant difference in the accuracy and reliability of your findings. Remember, the key to successful research is not just in collecting data, but in analyzing it thoughtfully and systematically to draw meaningful conclusions. So, dive in, explore, and contribute to the ever-growing body of knowledge with confidence. Happy researching!
At UoPeople, our blog writers are thinkers, researchers, and experts dedicated to curating articles relevant to our mission: making higher education accessible to everyone.
Related Articles
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 05 June 2024
A disease-associated gene desert directs macrophage inflammation through ETS2
- C. T. Stankey ORCID: orcid.org/0000-0001-5710-1716 1 , 2 , 3 na1 ,
- C. Bourges ORCID: orcid.org/0000-0001-8122-0475 1 na1 ,
- L. M. Haag ORCID: orcid.org/0000-0002-3754-5317 4 na1 ,
- T. Turner-Stokes 1 , 2 ,
- A. P. Piedade 1 ,
- C. Palmer-Jones 5 , 6 ,
- I. Papa ORCID: orcid.org/0000-0003-3167-7623 1 ,
- M. Silva dos Santos ORCID: orcid.org/0000-0003-2404-8490 7 ,
- Q. Zhang 8 ,
- A. J. Cameron ORCID: orcid.org/0000-0002-7065-9033 9 ,
- A. Legrini 9 ,
- T. Zhang 9 ,
- C. S. Wood 9 ,
- F. N. New ORCID: orcid.org/0000-0001-6213-4731 10 ,
- L. O. Randzavola 2 ,
- L. Speidel 11 , 12 ,
- A. C. Brown 13 ,
- A. Hall 14 , 15 ,
- F. Saffioti ORCID: orcid.org/0000-0001-7635-9931 6 , 14 ,
- E. C. Parkes 1 ,
- W. Edwards 16 ,
- H. Direskeneli 17 ,
- P. C. Grayson 18 ,
- L. Jiang 19 ,
- P. A. Merkel 20 , 21 ,
- G. Saruhan-Direskeneli ORCID: orcid.org/0000-0002-6903-7173 22 ,
- A. H. Sawalha ORCID: orcid.org/0000-0002-3884-962X 23 , 24 , 25 , 26 ,
- E. Tombetti 27 , 28 ,
- A. Quaglia 15 , 29 ,
- D. Thorburn 6 , 14 ,
- J. C. Knight ORCID: orcid.org/0000-0002-0377-5536 13 , 30 , 31 ,
- A. P. Rochford 5 , 6 ,
- C. D. Murray 5 , 6 ,
- P. Divakar 10 ,
- M. Green 32 ,
- E. Nye 32 ,
- J. I. MacRae ORCID: orcid.org/0000-0002-1464-8583 7 ,
- N. B. Jamieson ORCID: orcid.org/0000-0002-9552-4725 9 ,
- P. Skoglund 11 ,
- M. Z. Cader 16 , 33 ,
- C. Wallace ORCID: orcid.org/0000-0001-9755-1703 16 , 34 ,
- D. C. Thomas ORCID: orcid.org/0000-0002-9738-2329 16 , 33 &
- J. C. Lee ORCID: orcid.org/0000-0001-5711-9385 1 , 5 , 6
Nature volume 630 , pages 447–456 ( 2024 ) Cite this article
147k Accesses
1047 Altmetric
Metrics details
- Autoimmunity
- Functional genomics
- Immunogenetics
Increasing rates of autoimmune and inflammatory disease present a burgeoning threat to human health 1 . This is compounded by the limited efficacy of available treatments 1 and high failure rates during drug development 2 , highlighting an urgent need to better understand disease mechanisms. Here we show how functional genomics could address this challenge. By investigating an intergenic haplotype on chr21q22—which has been independently linked to inflammatory bowel disease, ankylosing spondylitis, primary sclerosing cholangitis and Takayasu’s arteritis 3 , 4 , 5 , 6 —we identify that the causal gene, ETS2 , is a central regulator of human inflammatory macrophages and delineate the shared disease mechanism that amplifies ETS2 expression. Genes regulated by ETS2 were prominently expressed in diseased tissues and more enriched for inflammatory bowel disease GWAS hits than most previously described pathways. Overexpressing ETS2 in resting macrophages reproduced the inflammatory state observed in chr21q22-associated diseases, with upregulation of multiple drug targets, including TNF and IL-23. Using a database of cellular signatures 7 , we identified drugs that might modulate this pathway and validated the potent anti-inflammatory activity of one class of small molecules in vitro and ex vivo. Together, this illustrates the power of functional genomics, applied directly in primary human cells, to identify immune-mediated disease mechanisms and potential therapeutic opportunities.
Similar content being viewed by others
The tidyomics ecosystem: enhancing omic data analyses
Genotype × environment interactions in gene regulation and complex traits

Genome-wide association studies
Nearly 5% of humans live with an autoimmune or inflammatory disease. These heterogeneous conditions, ranging from Crohn’s disease and ulcerative colitis (collectively inflammatory bowel disease (IBD)) to psoriasis and lupus, all require better therapies, but only 10% of drugs entering clinical development ever become approved treatments 2 . This high failure rate is mainly due to a lack of efficacy 8 and reflects our poor understanding of disease mechanisms. Genetics provides a unique opportunity to address this, with hundreds of loci now directly linked to the pathogenesis of immune-mediated diseases 9 . Indeed, drugs that target pathways implicated by genetics have a far higher chance of being effective 10 .
However, to fully realize the potential of genetics, knowledge of where risk variants lie must be translated into an understanding of how they drive disease 9 . Animal models can help with this, especially for coding variants in conserved genes 11 , 12 . Unfortunately, most risk variants do not lie in coding DNA, but in less-well-conserved, non-coding genomic regions. Resolving the biology at these loci is a formidable task, as the same DNA sequence can function differently depending on the cell type and/or external stimuli 9 . Most non-coding variants are thought to affect gene regulation 13 , but difficulties identifying causal genes, which may lie millions of bases away, and causal cell types, which may only express implicated genes under certain conditions, have hindered efforts to identify disease mechanisms. For example, although genome-wide association studies (GWASs) have identified over 240 IBD risk loci 3 , including several possible drug targets, fewer than 10 have been mechanistically resolved.
Molecular mechanisms at chr21q22
Some genetic variants predispose to multiple diseases, highlighting both their biological importance and an opportunity to study shared disease mechanisms. One notable example is an intergenic region on chromosome 21q22 (chr21q22), where the major allele haplotype predisposes to five inflammatory diseases 3 , 4 , 5 , 6 . Such regions, which were originally termed ‘gene deserts’ owing to their lack of coding genes, often contain GWAS hits but are poorly understood. To test for a shared disease mechanism, we performed co-localization analyses and confirmed that the genetic basis for every disease was the same, meaning that a common causal variant(s) and a shared molecular effect was responsible (Fig. 1a and Extended Data Fig. 1 ). As these heterogeneous diseases are all immune mediated, we reasoned that this locus must contain a distal enhancer that functioned in immune cells. By examining H3K27ac chromatin immunoprecipitation–sequencing (ChIP–seq) data, which marks active enhancers and promoters, we identified a monocyte/macrophage-specific enhancer within the locus (Fig. 1b ). Monocytes and macrophages have a key role in many immune-mediated diseases, producing cytokines that are often targeted therapeutically 14 .
a , Disease associations at chr21q22. The red points denote the IBD 99% credible set. Co-localization results for each disease versus IBD. PP.H3, posterior probability of independent causal variants; PP.H4, posterior probability of shared causal variant. b , Immune cell H3K27ac ChIP–seq at chr21q22. IBD GWAS results are shown. NK cells, natural killer cells. rpm, reads per million. c , The ETS2 eQTL in resting monocytes, with co-localization versus IBD association. Macrophage promoter-capture Hi-C (pcHi-C) data at the disease-associated locus. d , Experimental schematic for studying the chr21q22 locus in inflammatory (TPP) macrophages. e , ETS2 , BRWD1 and PSMG1 mRNA expression during TPP stimulation, measured using PrimeFlow RNA assays. Data are from one representative donor out of four. f , Relative ETS2 , BRWD1 and PSMG1 expression (mean fluorescence intensity (MFI)) in chr21q22-edited macrophages versus unedited cells. n = 4. Data are mean ± s.e.m. Statistical analysis was performed using two-way analysis of variance (ANOVA)). g , SuSiE fine-mapping posterior probabilities for IBD-associated SNPs at chr21q22 (99% credible set). h , Macrophage MPRA at chr21q22. Data are oligo coverage (top), enhancer activity (sliding-window analysis with significant enhancer activity highlighted; middle) and expression-modulating effects of SNPs within the enhancer (bottom). For the box plots, the centre line shows the median, the box limits show the interquartile range, and the whiskers represent the minimum and maximum values. n = 8. False-discovery rate (FDR)-adjusted P values were calculated using QuASAR-MPRA (two-sided). i , Inflammatory macrophage PU.1 ChIP–seq peaks at chr21q22. Bottom, magnification of the location of rs2836882 and the nearest predicted PU.1 motif. j , BaalChIP analysis of allele-specific PU.1 ChIP–seq binding at rs2836882 in two heterozygous macrophage datasets (data are mean ± 95% posterior distribution of allelic balance). Total counts shown as a pie chart. k , Allele-specific ATAC–seq reads at rs2836882 in monocytes from 16 heterozygous donors (including healthy controls and patients with ankylosing spondylitis). Statistical analysis was performed using two-sided Wilcoxon matched-pair tests. l , H3K27ac ChIP–seq data from risk (top) or non-risk (bottom) allele homozygotes at rs2836882. Data are shown from two out of four donors. FDR-corrected P values were calculated using MEDIPS (two-sided). The diagrams in d and e were created using BioRender.
Source Data
We next sought to identify the gene regulated by this enhancer. Although the associated locus lacks coding genes, there are several nearby candidates that have been highlighted in previous studies, including PSMG1 , BRWD1 and ETS2 (refs. 3 , 4 , 5 , 6 , 15 ) (Fig. 1a ). Using promoter-capture Hi-C and expression quantitative locus (eQTL) data from human monocytes ( Methods ), we found that the disease-associated locus physically interacts with the promoter of ETS2 —the most distant candidate gene (around 290 kb away)—and that the risk haplotype correlates with higher ETS2 expression (Fig. 1c ). Indeed, increased ETS2 expression in monocytes and macrophages, either at rest or after early exposure to bacteria, was found to have the same genetic basis as inflammatory disease risk (Extended Data Fig. 1c ). To directly confirm that ETS2 was causal, we used CRISPR–Cas9 to delete the 1.85 kb enhancer region in primary human monocytes before culturing these cells with inflammatory ligands, including TNF (a pro-inflammatory cytokine), prostaglandin E2 (a pro-inflammatory lipid) and Pam3CSK4 (a TLR1/2 agonist) (TPP model; Fig. 1d and Extended Data Fig. 2a–c ). This model was designed to mimic chronic inflammation 16 , and better resembles disease macrophages than classical IFNγ-driven or IL-4-driven models 17 (Extended Data Fig. 2 ). As flow cytometry antibodies were not available for the candidate genes, we used PrimeFlow to measure the dynamics of mRNA expression and detected increased levels of all three genes ( ETS2 , BRWD1 and PSMG1 ) after TPP stimulation of unedited monocytes (Fig. 1e ). Deletion of the chr21q22 enhancer did not affect BRWD1 or PSMG1 expression, but the upregulation of ETS2 was profoundly reduced (Fig. 1f ), confirming that this pleiotropic locus contains a distal ETS2 enhancer.
To identify the causal variant, we performed statistical fine-mapping in a large IBD GWAS 3 . Unfortunately, this did not resolve the association owing to high linkage disequilibrium between candidate single-nucleotide polymorphisms (SNPs) ( Methods and Fig. 1g ). We therefore used a functional approach to first delineate the active enhancers at the locus, and then assess whether any candidate SNPs might alter enhancer activity. This method, massively parallel reporter assay (MPRA), simultaneously tests enhancer activity in thousands of short DNA sequences by coupling each to a uniquely barcoded reporter gene 18 . Sequences that alter gene expression are identified by normalizing the barcode counts in mRNA, extracted from transfected cells, to their matching counts in the input DNA library. After adapting MPRA for primary macrophages ( Methods and Extended Data Fig. 3 ), we synthesized a pool of overlapping oligonucleotides to tile the 2 kb region containing all candidate SNPs, and included oligonucleotides with either risk or non-risk alleles for every variant. The resulting library was transfected into inflammatory macrophages from multiple donors, ensuring that a physiological repertoire of transcription factors could interact with the genomic sequences. Using a sliding-window analysis, we identified a single 442 bp focus of enhancer activity (chromosome 21: 40466236–40466677, hg19; Fig. 1h ) that contained three (out of seven) candidate SNPs. Two of these polymorphisms were transcriptionally inert, but the third (rs2836882) had the strongest expression-modulating effect of any candidate SNP, with the risk allele (G) increasing transcription, consistent with the ETS2 eQTL (Fig. 1h and Extended Data Fig. 1b ). This SNP was in the credible set of every co-localizing molecular trait, and lay within a macrophage PU.1 ChIP–seq peak (Fig. 1i ). PU.1 is a non-classical pioneer factor in myeloid cells 19 that can bind to DNA, initiate chromatin remodelling (thereby enabling other transcription factors to bind) and activate transcription 20 . To determine whether rs2836882 might affect PU.1 binding, we identified PU.1 ChIP–seq data from heterozygous macrophages and tested for allelic imbalances in binding. Despite not lying within a canonical PU.1 motif, strong allele-specific binding was detected, with over fourfold greater binding to the rs2836882 risk allele (Fig. 1i,j ). This was replicated by genotyping PU.1-bound DNA in macrophages from five heterozygous donors (Extended Data Fig. 4a–f ). Moreover, assay for transposase-accessible chromatin with sequencing (ATAC–seq) analysis of monocytes and macrophages from rs2836882 heterozygotes revealed allelic differences in chromatin accessibility that were consistent with differential binding of a pioneer factor (Fig. 1k and Extended Data Fig. 4g ).
To test for allele-specific enhancer activity at the endogenous locus, we performed H3K27ac ChIP–seq analysis of inflammatory macrophages from rs2836882 major and minor allele homozygotes. While most chr21q22 enhancer peaks were similar between these donors, the enhancer activity overlying rs2836882 was significantly stronger in major (risk) allele homozygotes (Fig. 1l and Extended Data Fig. 4h ), contributing to an approximate 2.5-fold increase in activity across the locus (Extended Data Fig. 4i ). Collectively, these data reveal a mechanism whereby the putative causal variant at chr21q22—identified by its functional effects in primary macrophages—promotes binding of a pioneer factor, enhances chromatin accessibility and increases activity of a distal ETS2 enhancer.
Macrophage inflammation requires ETS2
ETS2 is an ETS-family transcription factor and proto-oncogene 21 , but its exact role in human macrophages is unclear, with previous studies using either cell lines or complex mouse models and assessing a limited number of potential targets 22 , 23 , 24 , 25 , 26 . This has led to contradictory reports, with ETS2 being described as both necessary and redundant for macrophage development 27 , 28 , and both pro- and anti-inflammatory 22 , 23 , 24 , 25 , 26 . To clarify the role of ETS2 in human macrophages, and determine how dysregulated ETS2 expression might contribute to disease, we first used a CRISPR–Cas9-based loss-of-function approach (Fig. 2a ). To control for off-target effects, two gRNAs targeting different ETS2 exons were designed, validated and individually incorporated into Cas9 ribonucleoproteins for transfection into primary monocytes. These produced on-target editing in around 90% and 79% of cells, respectively, and effectively reduced ETS2 expression (Extended Data Fig. 2d–f ). Cell viability and macrophage marker expression were unaffected, suggesting that ETS2 was not required for macrophage survival or differentiation (Extended Data Fig. 2g,h ). By contrast, pro-inflammatory cytokine production, including IL-6, IL-8 and IL-1β, was markedly reduced after ETS2 disruption (Fig. 2b ), whereas IL-10—an anti-inflammatory cytokine—was less affected. TNF was not assessed as it had been added exogenously. We next investigated whether other macrophage functions were affected. Using fluorescently labelled particles that are detectable by flow cytometry, we found that phagocytosis was similarly impaired after ETS2 disruption (Fig. 2c ). We also tested extracellular reactive oxygen species (ROS) production—a major contributor to inflammatory tissue damage 29 . Disrupting ETS2 profoundly reduced the macrophage oxidative burst—most likely by decreasing expression of key NADPH oxidase components (Fig. 2d and Extended Data Fig. 5a ). Together, these data suggest that ETS2 is essential for multiple inflammatory functions in human macrophages.
a , Experimental schematic for studying ETS2 in inflammatory (TPP) macrophages. The diagram was created using BioRender. b , Cytokine secretion after ETS2 disruption. Heat map of relative cytokine levels from ETS2 -edited versus unedited macrophages. n = 8. c , Phagocytosis of fluorescently labelled zymosan particles by ETS2 -edited and unedited macrophages (non-targeting control (NTC)) (left). Data are from one representative donor out of seven. Right, the phagocytosis index (the product of the proportion and MFI of phagocytosing cells). n = 7. d , ROS production by ETS2 -edited and unedited macrophages. Data from one representative donor out of six (left). Right, NADPH oxidase component expression in ETS2 -edited and unedited macrophages (western blot densitometry). n = 7. Source gels are shown in Supplementary Fig. 1 . RLU, relative light units. e , RNA-seq analysis of differentially expressed genes in ETS2 -edited versus unedited TPP macrophages (limma with voom transformation, two-sided). n = 8. The horizontal line denotes the FDR-adjusted significance threshold. f , fGSEA of differentially expressed genes between ETS2 -edited and unedited TPP macrophages. The results of selected GO Biological Pathways are shown. The dot size denotes the unadjusted P value (two-sided), and the colour denotes normalized enrichment score (NES). g , The log 2 [fold change (FC)] of genes differentially expressed by chr21q22 enhancer deletion, plotted against their fold change after ETS2 editing. The percentages denote upregulated (red) and downregulated (blue) genes. The coloured points (blue or red) represent differentially expressed genes after ETS2 editing (FDR < 0.1, two-sided). For c and d , data are mean ± s.e.m. Statistical analysis was performed using two-sided Wilcoxon tests ( b – d ); * P < 0.05.
To understand the molecular basis for these effects, we performed RNA sequencing (RNA-seq) of ETS2 -edited and unedited inflammatory macrophages from multiple donors. Disrupting ETS2 led to widespread transcriptional changes, with reduced expression of many inflammatory genes (Fig. 2e ). These included cytokines (such as TNFSF10/TRAIL , TNFSF13 , IL1A and IL1B ), chemokines (such as CXCL3 , CXCL5 , CCL2 and CCL5 ), secreted effector molecules (such as S100A8 , S100A9 , MMP14 and MMP9 ), cell surface receptors (such as FCGR2A , FCGR2C and TREM1 ), pattern-recognition receptors (such as TLR2 , TLR6 and NOD2 ) and signalling molecules (such as MAP2K , GPR84 and NLRP3 ). To better characterize the pathways affected, we performed gene set enrichment analysis (fGSEA) using the Gene Ontology (GO) Biological Pathways dataset. This corroborated the functional deficits, with the most negatively enriched pathways (downregulated by ETS2 disruption) being related to macrophage activation, inflammatory cytokine production, phagocytosis and ROS production (Fig. 2f ). Genes involved in macrophage migration were also downregulated, but those relating to monocyte-to-macrophage differentiation were unaffected—consistent with ETS2 being required for inflammatory functions but not for monocyte-derived macrophage development. Fewer genes were upregulated after ETS2 disruption (Fig. 2e ), but positive enrichment was noted for aerobic respiration and oxidative phosphorylation (OXPHOS; Fig. 2f )—metabolic processes that are linked to anti-inflammatory phenotypes 30 . Notably, these transcriptional effects were not due to major changes in chromatin accessibility, although enhancer activity was generally reduced (Extended Data Fig. 2j,k ). As expected, deletion of the chr21q22 enhancer phenocopied both the transcriptional and functional effects of disrupting ETS2 (Fig. 2g and Extended Data Fig. 5a–e ). Collectively, these data identify an essential role for ETS2 in macrophage inflammatory responses, which could explain why dysregulated ETS2 expression predisposes to disease. Indeed, differential expression of ETS2-regulated genes was observed in resting (M0) macrophages from patients with IBD stratified by rs2836882 genotype (matched for age, sex, therapy and disease activity) (Extended Data Fig. 5f ).
ETS2 coordinates macrophage inflammation
We next studied the effects of increasing ETS2 expression, as this is what drives disease risk. To do this, we optimized a method for controlled overexpression of target genes in primary macrophages through transfection of in vitro transcribed mRNA that was modified to minimize immunogenicity (Fig. 3a , Methods and Extended Data Fig. 3f ). Resting, non-activated macrophages were transfected with ETS2 mRNA or its reverse complement, thereby controlling for mRNA quantity, length and purine/pyrimidine composition (Fig. 3b ). After transfection, cells were exposed to low-dose lipopolysaccharide to initiate a low-grade inflammatory response that could potentially be amplified (Fig. 3a ). We found that overexpressing ETS2 increased pro-inflammatory cytokine secretion, while IL-10 was again less affected (Extended Data Fig. 3g ). To better characterize this response, we performed RNA-seq and re-examined the inflammatory pathways that required ETS2 . Notably, all of these pathways—including macrophage activation, cytokine production, ROS production, phagocytosis and migration—were induced in a dose-dependent manner by ETS2 overexpression, with greater enrichment of every pathway when more ETS2 mRNA was transfected (Fig. 3c ). This shows that ETS2 is both necessary and sufficient for inflammatory responses in human macrophages, consistent with being a central regulator of effector functions, with dysregulation directly linked to disease.
a , Experimental schematic for studying the effects of ETS2 overexpression. The diagram was created using BioRender. b , ETS2 mRNA levels in transfected ( n = 8) or untransfected (from a separate experiment) macrophages. Data are mean ± s.e.m. CPM, counts per million. c , fGSEA analysis of differentially expressed genes between ETS2 -overexpressing and control macrophages. Results shown for pathways downregulated by ETS2 disruption. The dot size denotes the unadjusted P value (two-sided), the colour denotes NES and the border colour denotes the quantity of transfected mRNA. d , fGSEA analysis of a Crohn’s disease intestinal macrophage signature in ETS2 -overexpressing macrophages (versus control). FDR P -value, two-sided (top). Heat map of the relative expression of leading-edge genes after ETS2 overexpression (500 ng mRNA; bottom). e , Enrichment of macrophage signatures from patients with the indicated diseases in ETS2 -overexpressing macrophages (versus control). The colour denotes the disease category, the numbers denote the NES and the dashed line denotes FDR = 0.05. The Crohn’s disease signature is from a different study to that shown in d . AS, ankylosing spondylitis. f , SNPsea analysis of genes tagged by 241 IBD SNPs within ETS2 -regulated genes (red) and known IBD pathways (black). Significant pathways (Bonferroni-corrected P < 0.05) are indicated by hash symbols (#).
ETS2 has a key pathogenic role in IBD
To test whether ETS2 contributes to macrophage phenotypes in disease, we compared the effects of overexpressing ETS2 in resting macrophages with a single-cell RNA-seq (scRNA-seq) signature from intestinal macrophages in Crohn’s disease 31 . ETS2 overexpression induced a transcriptional state that closely resembled disease macrophages, with core (leading edge) enrichment of most signature genes, including several therapeutic targets (Fig. 3d ). Similar enrichment was observed with myeloid signatures from other chr21q22-associated diseases and, to a lesser extent, from active bacterial infection, but not for signatures from influenza and tumour macrophages, suggesting that ETS2 was not simply inducing generic activation (Fig. 3e ).
Given the central role of ETS2 in inflammatory macrophages and the importance of these cells in disease, we hypothesized that other genetic associations would also implicate this pathway. A major goal of GWAS was to identify disease pathways, but this has proven to be challenging due to a paucity of confidently identified causal genes and variants 9 . To determine whether the macrophage ETS2 pathway was enriched for disease genetics, we focused on IBD as this has more GWAS hits than any other chr21q22-associated disease. Encouragingly, a network of 33 IBD-associated genes in intestinal mucosa was previously found to be enriched for predicted ETS2 motifs 32 . Examining the genes that were consistently downregulated in ETS2 -edited macrophages (adjusted P ( P adj ) < 0.05 for both gRNAs), we identified over 20 IBD-risk-associated genes, including many thought to be causal at their respective loci 3 , 33 (Extended Data Table 1 ). These included genes that are known to affect macrophage biology (such as SP140 , LACC1 , CCL2 , CARD9 , CXCL5 , TLR4 , SLAMF8 and FCGR2A ) and some that are highly expressed in macrophages but not linked to specific pathways (such as ADCY7 , PTPRC , TAGAP , PTAFR and PDLIM5 ). A polygenic risk score comprising these variants associated with features of more severe IBD across 18,249 patients, including earlier disease onset, increased the need for surgery, and stricturing or fistulating complications in Crohn’s disease (Extended Data Fig. 6a–h ). To better test the enrichment of IBD GWAS hits in ETS2-mediated inflammation, and compare this with known disease pathways, we used SNPsea 34 —a method to identify pathways affected by disease loci. In total, 241 IBD loci were tested for enrichment in 7,658 GO Biological Pathways and 2 overlapping lists of ETS2-regulated genes (either those downregulated by ETS2 disruption or upregulated by ETS2 overexpression). Statistical significance was computed using 5 million matched null SNP sets, and pathways implicated by IBD genetics were extracted for comparison. Notably, IBD-associated SNPs were more significantly enriched in the macrophage ETS2 pathway than in many IBD pathways, with not a single null SNP set being more enriched in either ETS2-regulated gene list (Fig. 3f and Extended Data Fig. 6i ). SNPs associated with primary sclerosing cholangitis (PSC), ankylosing spondylitis and Takayasu’s arteritis were also enriched in ETS2-target genes (Extended Data Fig. 6j ). Collectively, this suggests that macrophage ETS2 signalling has a central role in multiple inflammatory diseases.
ETS2 has distinct inflammatory effects
We next investigated how ETS2 might control such diverse macrophage functions. Studying ETS2 biology is challenging because no ChIP-grade antibodies exist, precluding direct identification of its transcriptional targets. We therefore first used a guilt-by-association approach to identify genes that were co-expressed with ETS2 across 67 human macrophage activation conditions (comprising 28 stimuli and various durations of exposure) 16 . This identified PFKFB3 —encoding the rate-limiting enzyme of glycolysis—as the most highly co-expressed gene, with HIF1A also highly co-expressed (Fig. 4a ). Together, these genes facilitate a ‘glycolytic switch’ that is required for myeloid inflammatory responses 35 . We therefore hypothesized that ETS2 might control inflammation through metabolic reprogramming—a possibility supported by OXPHOS genes being negatively correlated with ETS2 (Fig. 4a ) and upregulated after ETS2 disruption (Fig. 2f ). To assess the metabolic consequences of disrupting ETS2 , we quantified label incorporation from 13 C-glucose in edited and unedited TPP macrophages using gas chromatography coupled with mass spectrometry (GC–MS). Widespread modest reductions in labelled and total glucose metabolites were detected after ETS2 disruption (Fig. 4b and Extended Data Fig. 7a–c ). This affected both glycolytic and tricarboxylic acid (TCA) cycle metabolites, with significant reductions in lactate, a hallmark of anaerobic glycolysis, and succinate, a key inflammatory metabolite 36 . These results are consistent with glycolytic suppression, with reductions in TCA metabolites being due to reduced flux into TCA and increased consumption by mitochondrial OXPHOS 37 . To determine whether metabolic changes accounted for ETS2-mediated inflammatory effects, we treated ETS2 -edited macrophages with roxadustat—a HIF1α stabilizer that promotes glycolysis. This had the predicted effect on glycolysis and OXPHOS genes, but did not rescue the effects of ETS2 disruption, either transcriptionally or functionally (Fig. 4c and Extended Data Fig. 7d,e ). Thus, while disrupting ETS2 impairs macrophage glycometabolism, this does not fully explain the differences in inflammation.
a , Genes co-expressed with ETS2 across 67 monocyte/macrophage activation conditions. The dotted lines denote FDR-adjusted P < 0.05. b , The effect of ETS2 disruption on glucose metabolism. The colour denotes median log 2 -transformed fold change in label incorporation from 13 C-glucose in ETS2 -edited versus unedited cells. The bold black border denotes P < 0.05 (Wilcoxon matched-pairs, two-sided). n = 6. Sec., secreted. c , fGSEA analysis of differentially expressed genes between ETS2 -edited and unedited macrophages that were treated with roxadustat or vehicle. Results shown for pathways downregulated by ETS2 disruption. d , Enrichment heat maps of macrophage ETS2 CUT&RUN peaks (IDR cut-off 0.01, n = 2) in 4 kb peak-centred regions from ATAC–seq (accessible chromatin), H3K4me3 ChIP–seq (active promoters) and H3K27ac ChIP–seq (active regulatory elements). e , Functional annotations of ETS2-binding sites (using gene coordinates and TPP macrophage H3K27ac ChIP–seq data). f , ETS2 motif enrichment in CUT&RUN peaks (hypergeometric P value, two-sided). g , ETS2 binding, chromatin accessibility (ATAC–seq) and regulatory activity (H3K27ac) at selected loci. h , Intersections between genes with ETS2 peaks in their core promoters or cis -regulatory elements and genes upregulated (Up) or downregulated (Dn) after ETS2 editing (KO) or overexpression (OE). The vertical bars denote the size of overlap for lists indicated by connected dots in the bottom panel. The horizontal bars denote the percentage of gene list within intersections. i , ETS2 binding, PU.1 binding, chromatin accessibility and enhancer activity at chr21q22. Predicted ETS2-binding sites (red) and PU.1-binding sites (purple) shown below. The dashed line is positioned at rs2836882.
We therefore revisited whether we could directly identify ETS2-target genes. As ChIP–seq involves steps that can alter protein epitopes and prevent antibody binding (such as fixation) we tested whether any anti-ETS2 antibodies might work for cleavage under targets and release using nuclease (CUT&RUN), which does not require these steps. One antibody identified multiple significantly enriched genomic regions (peaks), of which 6,560 were reproducibly detected across two biological replicates with acceptable quality metrics 38 (Fig. 4d ). These peaks were mostly located in active regulatory regions (90% in promoters or enhancers; Fig. 4d,e ) and were highly enriched for both a canonical ETS2 motif (4.02-fold versus global controls; Fig. 4f ) and for motifs of known ETS2 interactors, including FOS, JUN and NF-κB 39 (Extended Data Fig. 7f ). After combining the biological replicates to improve peak detection, we identified ETS2 binding at genes involved in multiple inflammatory functions, including NCF4 (ROS production), NLRP3 (inflammasome activation) and TLR4 (bacterial pattern recognition) (Fig. 4g ). Overall, 48.3% (754 out of 1,560) of genes dysregulated after ETS2 disruption and 50.3% (1,078 out of 2,153) of genes dysregulated after ETS2 overexpression contained an ETS2-binding peak within their core promoter or cis -regulatory elements (Fig. 4h ). Notably, ETS2 targets included HIF1A , PFKFB3 and other glycolytic genes (such as GPI , HK2 and HK3 ), consistent with the observed metabolic changes being directly induced as part of this complex inflammatory programme. Notably, we also detected ETS2 binding at the chr21q22 enhancer (Fig. 4i ). This is consistent with reports that PU.1 and ETS2 can interact synergistically 40 , and suggests that ETS2 might contribute to the activity of its own enhancer. Indeed, manipulating ETS2 expression altered enhancer activity in a manner consistent with positive autoregulation (Extended Data Fig. 7g–i ). Together, these data implicate ETS2 as a central regulator of monocyte and macrophage inflammatory responses that is able to direct a multifaceted effector programme and create a metabolic environment that is permissive for inflammation.
Targeting the ETS2 pathway in disease
To assess how ETS2 affects macrophage heterogeneity in diseased tissue, and whether this could be targeted therapeutically, we examined intestinal scRNA-seq data from patients with Crohn’s disease and healthy control individuals 41 . Within myeloid cells, seven clusters were detected and identified using established markers and/or previous literature (Fig. 5a,b ). Inflammatory macrophages (cluster 1, expressing CD209, CCL4, IL1B and FCGR3A) and inflammatory monocytes (cluster 2, expressing S100A8/A9, TREM1, CD14 and MMP9) were expanded in disease, as previously described 42 , and expressed ETS2 and ETS2-regulated genes more highly than other clusters, including tissue-resident macrophages (cluster 0, expressing C1QA, C1QB, FTL and CD63) and conventional dendritic cells (cluster 5, expressing CLEC9A, CADM1 and XCR1) (Fig. 5a,b and Extended Data Fig. 8a ). Using spatial transcriptomics, a similar increase in inflammatory macrophages was observed in PSC liver tissue, with these cells being closely apposed to cholangiocytes—the main target of pathology (Fig. 5c–e ). Notably, expression of ETS2-regulated genes was higher the closer macrophages were to cholangiocytes (Fig. 5f and Extended Data Fig. 8b ). Indeed, using bulk RNA-seq data, we found that the transcriptional footprint of ETS2 was detectable in affected tissues from multiple chr21q22-associated diseases (Extended Data Fig. 8c ).
a , Myeloid cell clusters in intestinal scRNA-seq from Crohn’s disease and health (top). Middle, scaled expression of ETS2-regulated genes (downregulated by ETS2 disruption). Bottom, the source of cells (disease or health). b , Scaled expression of selected genes. c , Spatial transcriptomics of PSC and healthy liver. n = 4. The images show representative fields of view (0.51 mm × 0.51 mm) with cell segmentation and semisupervised clustering. The main key (left and middle below images) denotes InSituType cell types; clusters a–e (far right key) are unannotated cell populations. Hep., hepatocyte; LSECs, liver sinusoidal endothelial cells; non-inflamm. macs, non-inflammatory macrophages. d , The number of macrophages within the indicated distances of cholangiocytes. e , The distance from cholangiocytes to the nearest macrophage. Data are shown as Tukey box and whisker plots. Statistical analysis was performed using two-tailed Mann–Whitney U -tests. Data in d and e are from 10,532 PSC and 13,322 control cholangiocytes. f , Scaled expression of ETS2-regulated genes in 21,067 PSC macrophages at defined distances from cholangiocytes (excluding genes used to define macrophage subsets). g , Classes of drugs that phenocopy ETS2 disruption (from the NIH LINCS database). h , fGSEA results for NIH LINCS drug signatures. Significant MEK inhibitor signatures are coloured by molecule. i , The log 2 [fold change] of differentially expressed genes after chr21q22 enhancer deletion, plotted against their fold change after MEK inhibition. The percentages indicate the proportion of upregulated (red) and downregulated genes (blue). The coloured points (blue or red) were differentially expressed after MEK inhibition (FDR < 0.1). j , fGSEA of differentially expressed genes between MEK-inhibitor-treated and control TPP macrophages. Results are shown for pathways downregulated by ETS2 disruption. The dot size denotes the unadjusted P value (two-sided) and the colour denotes the NES. k , IBD biopsy cytokine release with PD-0325901, infliximab or vehicle control. l , GSVA enrichment scores for chr21q22-downregulated genes in IBD biopsies after MEK inhibition. m , GSVA enrichment scores of a biopsy-derived molecular inflammation score (bMIS). Data are mean ± 95% CI ( f and l ) and mean ± s.e.m. ( k and m ). Statistical analysis was performed using two-sided paired t -tests. n = 10 ( k ), n = 9 ( l ). ** P < 0.01, *** P < 0.001, **** P < 0.0001.
We next examined whether this pathway could be targeted pharmacologically. Specific ETS2 inhibitors do not exist and structural analyses indicate that there is no obvious allosteric inhibitory mechanism 43 . We therefore used the NIH LINCS database to identify drugs that might modulate ETS2 activity 7 . This contains over 30,000 differentially expressed gene lists from cell lines exposed to around 6,000 small molecules. Using fGSEA, 906 signatures mimicked the effect of disrupting ETS2 ( P adj < 0.05), including several approved IBD therapies. The largest class of drugs was MEK inhibitors (Fig. 5g ), which are licensed for non-inflammatory human diseases (such as neurofibromatosis). This result was not due to a single compound, but rather a class effect with multiple MEK1/2 inhibitors downregulating ETS2-target genes (Fig. 5h ). This made biological sense, as MEK1/2, together with several other targets identified, are known regulators of ETS-family transcription factors (Fig. 5g ). Some of these compounds have shown benefit in animal colitis models 44 , although this is often a poor indicator of clinical efficacy, as several IBD treatments are ineffective in mice and many compounds that improve mouse models are ineffective in humans 45 . To test whether MEK inhibition abrogates ETS2-driven inflammation in human macrophages, we treated TPP macrophages with PD-0325901, a selective non-ATP competitive MEK inhibitor. Potent anti-inflammatory activity was observed that phenocopied the effects of disrupting ETS2 or the chr21q22 enhancer (Fig. 5i,j and Extended Data Fig. 9a–c ). To further assess the therapeutic potential, we cultured intestinal biopsies from active, untreated IBD with either a MEK inhibitor or a negative or positive control ( Methods ). MEK inhibition reduced inflammatory cytokine release to similar levels as infliximab (an anti-TNF antibody that is widely used for IBD; Fig. 5k ). Moreover, ETS2-regulated gene expression was reduced (Fig. 5l and Extended Data Fig. 9d ) and there was improvement in a transcriptional inflammation score 46 (Fig. 5m ). Together, these data show that targeting an upstream regulator of ETS2 can abrogate pathological inflammation in a chr21q22-associated disease, and may be useful therapeutically.
Arguably the greatest challenge in modern genetics is to translate the success of GWAS into a better understanding of disease. Here, by studying a pleiotropic disease locus, we identify a central regulator of human macrophage inflammation and a pathogenic pathway that is potentially druggable. These findings also provide clues to the gene–environment interactions at this locus, highlighting a potential role for ETS2 in macrophage responses to bacteria. This would provide a balancing selection pressure that might explain why the risk allele remains so common (frequency of around 75% in Europeans and >90% in Africans) despite first being detected in archaic humans over 500,000 years ago (Extended Data Fig. 10 ).
Although ETS2 was reported to have pro-inflammatory effects on individual genes 24 , 25 , the full extent of its inflammatory programme—with effects on ROS production, phagocytosis, glycometabolism and macrophage activation—was unclear. Moreover, without direct proof of ETS2 targets, nor studies in primary human cells, it was difficult to reconcile reports of anti-inflammatory effects at other genes 23 , 26 . By systematically characterizing the effects of ETS2 disruption and overexpression in human macrophages, we identify an essential role in inflammation, delineate the mechanisms involved and show how ETS2 can induce pathogenic macrophage phenotypes. Increased ETS2 expression may also contribute to other human pathology. For example, Down’s syndrome (trisomy 21) was recently described as a cytokinopathy 47 , with basal increases in multiple inflammatory cytokines, including several ETS2 targets (such as IL-1β, TNF and IL-6). Whether the additional copy of ETS2 contributes to this phenotype is unknown, but warrants further study.
Blocking individual cytokines is a common treatment strategy in inflammatory disease 14 , but emerging evidence suggests that targeting several cytokines at once may be a better approach 48 . Blocking ETS2 signalling through MEK1/2 inhibition affects multiple cytokines, including TNF and IL-23, which are targets of existing therapies, and IL-1β, which is linked to treatment resistance 49 and not directly modulated by other small molecules (such as JAK inhibitors). However, long-term MEK inhibitor use may not be ideal owing to the physiological roles of MEK in other tissues, with multiple side-effects having been reported 50 . Targeting ETS2 directly—for example, through PROTACs—or selectively delivering MEK inhibitors to macrophages through antibody–drug conjugates could overcome this toxicity, and provide a safer means of blocking ETS2-driven inflammation.
In summary, using an intergenic GWAS hit as a starting point, we have identified a druggable pathway that is both necessary and sufficient for human macrophage inflammation. Moreover, we show how genetic dysregulation of this pathway—through perturbation of pioneer factor binding at a critical long-range enhancer—predisposes to multiple diseases. This highlights the considerable, yet largely untapped, opportunity to resolve disease biology from non-coding genetic associations.
Analysis of existing data relating to chr21q22
IBD GWAS summary statistics 3 were used to perform multiple causal variant fine-mapping using susieR 51 , with reference minor allele and LD information calculated from 503 European samples from 1000 Genomes phase 3 (ref. 52 ). All R analyses used v.4.2.1. Palindromic SNPs (A/T or C/G) and any SNPs that did not match by position or alleles were pruned before imputation using the ssimp equations reimplemented in R. This did not affect any candidate SNP at chr21q22. SuSiE fine-mapping results were obtained for ETS2 (identifier ENSG00000157557 or ILMN_1720158) in monocyte/macrophage datasets from the eQTL Catalogue 53 . Co-localization analyses were performed comparing the chr21q22 IBD association with summary statistics from other chr21q22-associated diseases 3 , 4 , 5 , 6 and monocyte/macrophage eQTLs 54 , 55 , 56 , 57 , 58 to determine whether there was a shared genetic basis for these different associations. This was performed using coloc (v.5.2.0) 59 using a posterior probability of H4 (PP.H4.abf) > 0.5 to call co-localization.
Raw H3K27ac ChIP–seq data from primary human immune cells were downloaded from Gene Expression Omnibus (GEO series GSE18927 and GSE96014 ) and processed as described previously 60 (code provided in the ‘Code availability’ section).
Processed promoter-capture Hi-C data 61 from 17 primary immune cell types were downloaded from OSF ( https://osf.io/u8tzp ) and cell type CHiCAGO scores for chr21q22-interacting regions were extracted.
Monocyte-derived macrophage differentiation
Leukocyte cones from healthy donors were obtained from NHS Blood and Transplant (Cambridge Blood Donor Centre, Colindale Blood Centre or Tooting Blood Donor Centre). Peripheral blood mononuclear cells (PBMCs) were isolated by density centrifugation (Histopaque 1077, Sigma-Aldrich) and monocytes were positively selected using CD14 Microbeads (Miltenyi Biotec). Macrophage differentiation was performed either using conditions that model chronic inflammation (TPP) 16 : 3 days GM-CSF (50 ng ml −1 , Peprotech) followed by 3 days GM-CSF, TNF (50 ng ml −1 , Peprotech), PGE 2 (1 μg ml −1 , Sigma-Aldrich) and Pam 3 CSK4 (1 μg ml −1 , Invivogen); or, to produce resting (M0) macrophages: 6 days M-CSF (50 ng ml −1 , Peprotech). All cultures were performed at 37 °C under 5% CO 2 in antibiotic-free RPMI1640 medium containing 10% FBS, GlutaMax and MEM non-essential amino acids (all Thermo Fisher Scientific). Cells were detached using Accutase (BioLegend).
Identifying a model of chronic inflammatory macrophages
Human monocyte/macrophage gene expression data files ( n = 314) relating to 28 different stimuli with multiple durations of exposure (collectively comprising 67 different activation conditions) were downloaded from the GEO ( GSE47189 ) and quantile normalized. Data from biological replicates were summarized to the median value for every gene. Gene set variation analysis 62 (using the GSVA package in R) was performed to identify the activation condition that most closely resembled CD14 + monocytes/macrophages from active IBD using disease-associated lists of differentially expressed genes 63 .
CRISPR–Cas9 editing of primary human monocytes
gRNA sequences were designed using CRISPick and synthesized by IDT (Supplementary Table 3 ). Alt-R CRISPR–Cas9 negative control crRNA 1 (IDT) was used as a non-targeting control. Cas9–gRNA ribonucleoproteins were assembled as described previously 60 and nucleofected into 5 × 10 6 monocytes in 100 μl nucleofection buffer (Human Monocyte Nucleofection Kit, Lonza) using a Nucleofector 2b (Lonza, program Y-001). After nucleofection, monocytes were immediately transferred into 5 ml of prewarmed culture medium in a six-well plate, and differentiated into macrophages under TPP conditions. The editing efficiency was quantified by PCR amplification of the target region in extracted DNA. All primer sequences are provided in Supplementary Table 3 . The editing efficiency at the chr21q22 locus was measured by quantification of amplified fragments (2100 Bioanalyzer, Agilent) as previously described 60 . The editing efficiency for individual gRNAs was assessed using the Inference of CRISPR Edits tool 64 (ICE, Synthego).
PrimeFlow RNA assay
RNA abundance was quantified by PrimeFlow (Thermo Fisher Scientific) in chr21q22-edited and unedited (NTC) cells on days 0, 3, 4, 5 and 6 of TPP differentiation. Target probes specific for ETS2 (Alexa Fluor 647), BRWD1 (Alexa Fluor 568) and PSMG1 (Alexa Fluor 568) were used according to the manufacturer’s instructions. Data were collected using FACS Diva software and analysed using FlowJo v10 (BD Biosciences).
Overlapping oligonucleotides containing 114 nucleotides of genomic sequence were designed to tile the region containing chr21q22 candidate SNPs (99% credible set) at 50 bp intervals. Six technical replicates were designed for every genomic sequence, each tagged by a unique 11-nucleotide barcode. Additional oligonucleotides were included to test the expression-modulating effect of every candidate SNP in the 99% credible set. Allelic constructs were designed as described previously 60 and tagged by 30 unique 11-nucleotide barcodes. Positive and negative controls were included as described previously 60 . 170-nucleotide oligonucleotides were synthesized as part of a larger MPRA pool (Twist Biosciences) containing the 16-nucleotide universal primer site ACTGGCCGCTTCACTG, 114-nucleotide variable genomic sequence, KpnI and XbaI restriction sites (TGGACCTCTAGA), an 11-nucleotide barcode and the 17-nucleotide universal primer site AGATCGGAAGAGCGTCG. Cloning into the MPRA vector was performed as described previously 60 . A suitable promoter for the MPRA vector (RSV) was identified by testing promoter activities in TPP macrophages. The MPRA vector library was nucleofected into TPP macrophages (5 µg vector into 5 × 10 6 cells) in 100 μl nucleofection buffer (Human Macrophage Nucleofection Kit, Lonza) using a Nucleofector 2b (program Y-011). To ensure adequate barcode representation, a minimum of 2 × 10 7 cells was nucleofected for every donor ( n = 8). After 24 h, RNA was extracted and sequencing libraries were made from mRNA or DNA input vector as described previously 60 . Libraries were sequenced on the Illumina HiSeq2500 high-output flow-cell (50 bp, single-end reads). Data were demultiplexed and converted to FASTQ files using bcl2fastq and preprocessed as previously described using FastQC 60 . To identify regions of enhancer activity, a paired t -test was first performed to identify genomic sequences that enhanced transcription and a sliding-window analysis (300 bp window) was then performed using the les package in R. Expression-modulating variants were identified using QuASAR-MPRA 65 , as described previously 60 .
Publicly available PU.1 ChIP–seq datasets from human macrophages were downloaded from GEO, and BAM files were examined (IGV genome browser) to identify heterozygous samples (that is, files containing both A and G allele reads at chr21:40466570; hg19). Two suitable samples were identified ( GSM1681423 and GSM1681429 ) and used for a Bayesian analysis of allelic imbalances in PU.1 binding (implemented in the BaalChIP package 66 in R) with correction for biases introduced by overdispersion and biases towards the reference allele.
Allele-specific PU.1 ChIP genotyping
A 100 ml blood sample was taken from five healthy rs2836882 heterozygotes (assessed by Taqman genotyping; Thermo Fisher Scientific). All of the participants provided written informed consent. Ethical approval was provided by the London–Brent Regional Ethics Committee (21/LO/0682). Monocytes were isolated from PBMCs using CD14 Microbeads (Miltenyi Biotec) and differentiated into inflammatory macrophages using TPP conditions 16 . After differentiation, macrophages were detached and cross-linked for 10 min in fresh medium containing 1% formaldehyde. Cross-linking was quenched with glycine (final concentration 0.125 M, 5 min). Nucleus preparation and shearing were performed as described previously 60 with 10 cycles sonication (30 s on/30 s off, Bioruptor Pico, Diagenode). PU.1 was immunoprecipitated overnight at 4 °C using a polyclonal anti-PU.1 antibody (1:25; Cell Signaling) using the SimpleChIP Plus kit (Cell Signaling). The ratio of rs2836882 alleles in the PU.1-bound DNA was quantified in duplicate by TaqMan genotyping (assay C 2601507_20). A standard curve was generated using fixed ratios of geneblocks containing either the risk or non-risk allele (200-nucleotide genomic sequence centred on rs2836882; Genewiz).
PU.1 MPRA ChIP–seq
The MPRA vector library was transfected into TPP macrophages from six healthy donors. Assessment of PU.1 binding to SNP alleles was performed as described previously 60 , with minimal sonication (to remove contaminants without chromatin shearing). Immunoprecipitation was performed as described above. Sequencing libraries were prepared as for MPRA and sequenced on the MiSeq system (50 bp, single-end reads).
ATAC–seq analysis
ATAC–seq in ETS2 -edited and unedited TPP macrophages was performed using the Omni-ATAC protocol 67 with the following modifications: the cell number was increased to 75,000 cells; the cell lysis time was increased to 5 min; the volume of Tn5 transposase in the transposition mixture was doubled; and the duration of the transposition step was extended to 40 min. Amplified libraries were cleaned using AMPure XP beads (Beckman Coulter) and sequenced on the NovaSeq6000 system (100 bp paired-end reads). Data were processed as described previously 68 . Differential ATAC–seq analysis was performed as described previously using edgeR and TMM normalization 69 . Allele-specific ATAC–seq analysis was performed in 16 heterozygous monocyte datasets from healthy controls and patients with ankylosing spondylitis 70 and in 2 deeply sequenced heterozygous TPP macrophage samples. For these analyses, sequencing reads at rs2836882 were extracted from preprocessed data using splitSNP ( https://github.com/astatham/splitSNP ) (see the ‘Code availability’ section).
H3K27ac ChIP–seq
H3K27ac ChIP–seq was performed as described previously 60 using an anti-H3K27ac antibody (1:250, Abcam) or an isotype control (1:500, rabbit IgG, Abcam). Sequencing libraries from TPP macrophages from major and minor allele homozygotes at rs2836882 (identified through the NIHR BioResource, n = 4) were sequenced on the HiSeq4000 system (50 bp, single-end reads). Sequencing libraries from ETS2 -edited and unedited TPP macrophages ( n = 3) or resting M0 macrophages overexpressing ETS2 or control mRNA ( n = 3) were sequenced on the NovaSeq6000 system (100 bp, paired-end reads). Raw data were processed, quality controlled and analysed as described previously using the Burrows-Wheeler Aligner 60 . Unpaired differential ChIP–seq analysis, to compare rs2836882 genotypes, was performed using MEDIPS 71 by dividing the 560 kb region around rs2836882 (chr21:40150000–40710000, hg19) into 5 kb bins. Paired differential ChIP–seq analyses, to assess the effect of perturbing ETS2 expression on enhancer activity, were performed using edgeR with TMM normalization 69 , 72 (with donor as covariate). Genome-wide analyses used consensus MACS2 peaks. Superenhancer activity was evaluated using Rank-Ordering of Super-Enhancers (ROSE). Chr21q22-based analyses used the enhancer coordinates that exhibited allele-specific activity (chr21:40465000–40470000, hg19). Code is provided for all data analysis (see the ‘Code availability’ section).
Assays of macrophage effector functions
Flow cytometry.
Expression of myeloid markers was assessed using flow cytometry (BD LSRFortessa X-20) with the following panel: CD11b PE/Dazzle 594 (BioLegend), CD14 evolve605 (Thermo Fisher Scientific), CD16 PerCP (BioLegend), CD68 FITC (BioLegend), Live/Dead Fixable Aqua Dead Cell Stain (Thermo Fisher Scientific) and Fc Receptor Blocking Reagent (Miltenyi). All antibodies were used at a dilution of 1:40; Live/Dead stained was used at 1:400 dilution. Data were collected using FACS Diva and analysed using FlowJo v.10 (BD Biosciences).
Cytokine quantification
Supernatants were collected on day 6 of TPP macrophage culture and frozen. Cytokine concentrations were quantified in duplicate by electrochemiluminescence using assays (Meso Scale Diagnostics, DISCOVERY WORKBENCH v.4.0).
Phagocytosis
Phagocytosis was assessed using fluorescently labelled Zymosan particles (Green Zymosan, Abcam) according to the manufacturer’s instructions. Cells were seeded at 10 5 cells per well in 96-well round-bottom plates. Cytochalasin D (10 μg ml −1 , Thermo Fisher Scientific) was used as a negative control. Phagocytosis was quantified by flow cytometry, and a phagocytosis index was calculated (the proportion of positive cells multiplied by their mean fluorescence intensity).
Extracellular ROS production
Extracellular ROS production was quantified using the Diogenes Enhanced Superoxide Detection Kit (National Diagnostics) according to the manufacturer’s protocol. Cells were seeded at a density of 10 5 cells per well and prestimulated with PMA (200 ng ml −1 , Sigma-Aldrich).
Western blotting
Western blotting was performed as described previously 73 using the following primary antibodies: mouse anti-gp91phox (1:2,000), mouse anti-p22phox (1:500; both Santa Cruz), rabbit anti-C17ORF62/EROS (1:1,000; Atlas), mouse anti-vinculin (Sigma-Aldrich). Loading controls were run on the same gel. Secondary antibodies were as follows: goat anti-rabbit IgG-horseradish or goat anti-mouse IgG-horseradish peroxidase (both 1:10,000; Jackson Immuno). Chemiluminescence was recorded on the ChemiDoc Touch imager (Bio-Rad) after incubation of the membrane with ECL (Thermo Fisher Scientific) or SuperSignal West Pico PLUS (Thermo Fisher Scientific) reagent. Densitometry analysis was performed using ImageJ.
RNA-seq analysis
RNA was isolated from macrophage lysates (AllPrep DNA/RNA Micro Kit, Qiagen) and sequencing libraries were prepared from 10 ng RNA using the SMARTer Stranded Total RNA-Seq Kit v2 Pico Input Mammalian (Takara) according to the manufacturer’s instructions. Libraries were sequenced on either the NextSeq 2000 (50 bp paired-end reads: CRISPR, roxadustat and PD-0325901 experiments) or NovaSeq 6000 (100 bp paired-end reads: overexpression experiments) system and preprocessed using MultiQC. Reads were trimmed using Trim Galore (Phred score 24) and filtered to remove reads <20 bp. Ribosomal reads (mapping to human ribosomal DNA complete repeating unit; GenBank: U13369 .1 ) were removed using BBSplit ( https://sourceforge.net/projects/bbmap/ ). Reads were aligned to the human genome (hg38) using HISAT2 (ref. 74 ) and converted to BAM files, sorted and indexed using SAMtools 75 . Gene read counts were obtained using the featureCounts program 76 from Rsubread using the GTF annotation file for GRCh38 (v.102). Differential expression analysis was performed in R using limma 77 with voom transformation and including donor as a covariate. Differential expression results are shown in Supplementary Tables 1 and 2 .
GSEA was performed using fGSEA 78 in R with differentially expressed gene lists ranked by t -statistic. Gene sets were obtained from GO Biological Pathways (MSigDB), experimentally derived based on differential expression analysis or sourced from published literature 31 , 42 , 70 , 79 , 80 , 81 , 82 , 83 , 84 , 85 , 86 . Specific details of disease macrophage signatures (Fig. 3f ) are provided as source data. GO pathways shown in Figs. 2 – 5 are as follows: GO:0002274, GO:0042116, GO:0097529, GO:0006909, GO:0071706, GO:0032732, GO:0032755, GO:0032757, GO:2000379, GO:0009060, GO:0006119 and GO:0045649. Statistical significance was calculated using the adaptive multilevel split Monte Carlo method.
IBD BioResource recall-by-genotype study
IBD patients who were rs2836882 major or minor allele homozygotes ( n = 11 of each) were identified through the NIHR IBD BioResource. Patients were matched for age, sex, treatment and disease activity, and all provided written informed consent. Ethical approval was provided by the London–Brent Regional Ethics Committee (21/LO/0682). A 50 ml blood sample was taken from all patients and M0 monocyte-derived macrophages were generated as described. After 6 days, cells were collected, lysed and RNA was extracted. Quantitative PCR analysis of a panel of ETS2-regulated genes was performed in triplicate after reverse transcription (SuperScript IV VILO, Thermo Fisher Scientific) using the Quantifast SYBR Green PCR kit (Qiagen) on the Roche LightCycler 480. Primer sequences are provided in Supplementary Table 3 and PPIA and RPLP0 were used as housekeeping genes. Expression values for each gene ( \({2}^{\Delta {c}_{T}}\) ) were scaled to a minimum 0 and maximum 1 to enable intergene comparison.
In vitro transcription
The cDNA sequence for ETS2 (NCBI Reference Sequence Database NM005239.5 ) preceded by a Kozak sequence was synthesized and cloned into a TOPO vector. This was linearized and a PCR amplicon generated, adding a T7 promoter and an AG initiation sequence (Phusion, NEB). A reverse complement (control) amplicon was also generated. These amplicons were used as templates for in vitro transcription using the HiScribe T7 mRNA Kit with CleanCap Reagent AG kit (NEB) according to the manufacturer’s instructions, but with substitution of N1-methyl-pseudouridine for uridine and methylcytidine for cytidine (both Stratech) to minimize non-specific cellular activation by the transfected mRNA. mRNA was purified using the MEGAclear Kit (Thermo Fisher Scientific) and polyadenylated using an Escherichia coli poly(A) polymerase (NEB) before further clean-up (MEGAclear), quantification and analysis of the product size (NorthernMax-Gly gel, Thermo Fisher Scientific). For optimizing overexpression conditions, GFP mRNA was produced using the same method. All primer sequences are provided in Supplementary Table 3 .
mRNA overexpression
Lipofectamine MessengerMAX (Thermo Fisher Scientific) was diluted in Opti-MEM (1:75 v/v), vortexed and incubated at room temperature for 10 min. IVT mRNA was then diluted in a fixed volume of Opti-MEM (112.5 µl per transfection), mixed with an equal volume of diluted Lipofectamine MessengerMAX and incubated for a further 5 min at room temperature. The transfection mix was then added dropwise to 2.5 × 10 6 M0 macrophages (precultured for 6 days in a six-well plate in antibiotic-free RPMI1640 macrophage medium containing M-CSF (50 ng ml −1 , Peprotech), with medium change on day 3). For GFP overexpression, cells were detached using Accutase 18 h after transfection and GFP expression was measured using flow cytometry. For ETS2 /control overexpression, either 250 ng or 500 ng mRNA was transfected and low-dose LPS (0.5 ng ml −1 ) was added 18 h after transfection, and cells were detached using Accutase 6 h later. Representative ETS2 expression in untransfected macrophages was obtained from previous data ( GSE193336 ). Differential H3K27ac ChIP–seq analysis in ETS2 -overexpressing macrophages was performed using 500 ng RNA transfection (see the ‘Code availability’ section).
Plink1.9 ( https://www.cog-genomics.org/plink/1.9/ ) was used to calculate a polygenic risk score (PRS) for patients in the IBD BioResource using 22 ETS2-regulated IBD-associated SNPs ( β coefficients from a previous study 3 ). Linear regression was used to compare PRSs with age at diagnosis, and logistic regression to estimate the effect of PRSs on IBD subphenotypes, including anti-TNF primary non-response (PNR), CD behaviour (B1 versus B2/B3), perianal disease and surgery. For variables with more than two levels (for example, CD location or UC location), ANOVA was used to investigate the relationship with PRS. For analyses of age at diagnosis, anti-TNF response and surgery, IBD diagnosis was included as a covariate.
Pathway analysis of 241 IBD-associated GWAS hits 3 was performed using SNPsea v.1.0.4 (ref. 34 ). In brief, linkage intervals were defined for every lead SNP based on the furthest correlated SNPs ( r 2 > 0.5 in 1000 Genomes, European population) and were extended to the nearest recombination hotspots with recombination rate > 3 cM per Mb. If no genes were present in this region, the linkage interval was extended up- and downstream by 500 kb (as long-range regulatory interactions usually occur within 1 Mb). Genes within linkage intervals were tested for enrichment within 7,660 pathways, comprising 7,658 GO Biological Pathways and two lists of ETS2-regulated genes (either those significantly downregulated after ETS2 disruption with gRNA1 or those significantly upregulated after ETS2 overexpression, based on a consensus list obtained from differential expression analysis including all samples and using donor and mRNA quantity as covariates). The analysis was performed using a single score mode: assuming that only one gene per linkage interval is associated with the pathway. A null distribution of scores for each pathway was performed by sampling identically sized random SNP sets matched on the number of linked genes (5,000,000 iterations). A permutation P value was calculated by comparing the score of the IBD-associated gene list with the null scores. An enrichment statistic was calculated using a standardized effect size for the IBD-associated score compared to the mean and s.e.m. of the null scores. Gene sets relating to the following IBD-associated pathways were extracted for comparison: NOD2 signalling (GO:0032495), integrin signalling (GO:0033627, GO:0033622), TNF signalling (GO:0033209, GO:0034612), intestinal epithelium (GO:0060729, GO:0030277), Th17 cells (GO:0072539, GO:0072538, GO:2000318), T cell activation (GO:0046631, GO:0002827), IL-10 signalling (GO:0032613, GO:0032733) and autophagy (GO:0061919, GO:0010506, GO:0010508, GO:1905037, GO:0010507). SNPs associated with PSC 5 , 87 , ankylosing spondylitis 4 , 87 , Takayasu arteritis 6 , 88 , 89 and schizophrenia 90 (as a negative control) were collated from the indicated studies and tested for enrichment in ETS2-regulated gene lists.
ETS2 co-expression
Genes co-expressed with ETS2 across 67 human monocyte/macrophage activation conditions (normalized data from GSE47189 ) were identified using the rcorr function in the Hmisc package in R.
13 C-glucose GC–MS
ETS2 -edited or unedited TPP macrophages were generated in triplicate for each donor and on day 6, the medium was removed, cells were washed with PBS, and new medium with labelled glucose was added. Labelled medium was as follows: RPMI1640 medium, no glucose (Thermo Fisher Scientific), 10% FBS (Thermo Fisher Scientific), GlutaMax (Thermo Fisher Scientific), 13 C-labelled glucose (Cambridge Isotype Laboratories). After 24 h, a timepoint selected from a time-course to establish steady-state conditions, the supernatants were snap-frozen and macrophages were detached by scraping. Macrophages were washed three times with ice-cold PBS, counted, resuspended in 600 µl ice-cold chloroform:methanol (2:1, v/v) and sonicated in a waterbath (3 times for 8 min). All of the extraction steps were performed at 4 °C as previously described 91 . The samples were analysed on the Agilent 7890B-7000C GC–MS system. Spitless injection (injection temperature of 270 °C) onto a DB-5MS (Agilent) was used, using helium as the carrier gas, in electron ionization mode. The initial oven temperature was 70 °C (2 min), followed by temperature gradients to 295 °C at 12.5 °C per min and to 320 °C at 25 °C per min (held for 3 min). The scan range was m / z 50–550. Data analysis was performed using in-house software MANIC (v.3.0), based on the software package GAVIN 92 . Label incorporation was calculated by subtracting the natural abundance of stable isotopes from the observed amounts. Total metabolite abundance was normalized to the internal standard (scyllo-inositol 91 ).
Roxadustat in TPP macrophages
ETS2- edited or unedited TPP macrophages were generated as described previously. On day 5 of culture, cells were detached (Accutase) and replated at a density of 10 5 cells per well in 96-well round-bottom plates in TPP medium containing roxadustat (FG-4592, 30 μM). After 12 h, cells were collected for functional assays and RNA-seq as described.
CUT&RUN
Precultured TPP macrophages were collected and processed immediately using the CUT&RUN Assay kit (Cell Signaling) according to the manufacturer’s instructions but omitting the use of ConA-coated beads. In brief, 5 × 10 5 cells per reaction were pelleted, washed and resuspended in antibody binding buffer. Cells were incubated with antibodies: anti-ETS2 (1:100, Thermo Fisher Scientific) or IgG control (1:20, Cell Signaling) for 2 h at 4 °C. After washing in digitonin buffer, cells were incubated with pA/G-MNase for 1 h at 4 °C. Cells were washed twice in digitonin buffer, resuspended in the same buffer and cooled for 5 min on ice. Calcium chloride was added to activate pA/G-MNase digestion (30 min, 4 °C) before the reaction was stopped and cells incubated at 37 °C for 10 min to release cleaved chromatin fragments. DNA was extracted from the supernatants using spin columns (Cell Signaling). Library preparation was performed using the NEBNext Ultra II DNA Library Prep Kit according to a protocol available at protocols.io ( https://doi.org/10.17504/protocols.io.bagaibse ). Size selection was performed using AMPure XP beads (Beckman Coulter) and the fragment size was assessed using the Agilent 2100 Bioanalyzer (High Sensitivity DNA kit). Indexed libraries were sequenced on the NovaSeq 6000 system (100 bp paired-end reads). Raw data were analysed using guidelines from the Henikoff laboratory 93 . In brief, paired-end reads were trimmed using Trim Galore and aligned to the human genome (GRCh37/hg19) using Bowtie2. BAM files were sorted, merged (technical and, where indicated, biological replicates), resorted and indexed using SAMtools. Picard was used to mark unmapped reads and SAMtools to remove these, re-sort and re-index. Bigwig files were created using the deepTools bamCoverage function. Processed data were initially analysed using the nf-core CUT&RUN pipeline v.3.0, using CPM normalization and default MACS2 parameters for peak calling. This analysis yielded acceptable quality metrics (including an average FRiP score of 0.23) but there was a high number of peaks with low fold enrichment (<4) over the control. More stringent parameters were therefore applied for peak calling (--qvalue 0.05 -f BAMPE --keep-dup all -B --nomodel) and we applied an irreproducible discovery rate (IDR; cut-off 0.001) to identify consistent peaks between replicates, implemented in the idr package in R (see the ‘Code availability’ section). Enrichment of binding motifs for ETS2 and other transcription factors expressed in TPP macrophages (cpm > 0.5) within consensus IDR peaks was calculated using TFmotifView 94 using global genomic controls. The overlap between consensus IDR peaks and the core promoter (−250bp to +35 bp from the transcription start site) and/or putative cis -regulatory elements of ETS2-regulated genes was assessed using differentially expressed gene lists after ETS2 disruption (gRNA1) or ETS2 overexpression (based on a consensus across mRNA doses, as described earlier). Putative cis -regulatory elements were defined as shared interactions (CHiCAGO score > 5) in monocyte and M0 and M1 macrophage samples from publicly available promoter-capture Hi-C data 61 . Predicted ETS2- and PU.1-binding sites were identified at the rs2836882 locus (chr21:40466150–40467450) using CisBP 95 (database 2.0, PWMs log odds motif model, default settings).
Intestinal scRNA-seq
Raw count data from colonic immune cells 41 (including healthy controls and Crohn’s disease) were downloaded from the Single Cell Portal ( https://singlecell.broadinstitute.org/single_cell ). Myeloid cell data were extracted for further analysis using the cell annotation provided. Raw data were preprocessed, normalized and variance-stabilized using Seurat (v.4) 96 . PCA and UMAP clustering was performed and clusters annotated using established markers and/or previous literature. Marker genes were identified using the FindAllMarkers function. Modular expression of ETS2-regulated genes (downregulated after ETS2 editing, gRNA1) was measured using the AddModuleScore function.
Spatial transcriptomics
Formalin-fixed paraffin-embedded sections (thickness, 5 μm) were cut from two PSC liver explants and two controls (healthy liver adjacent to tumour metastases), baked overnight at 60 °C and prepared for CosMx according to manufacturer’s instructions using 15 min target retrieval and 30 min protease digestion. Tissue samples were obtained through Tissue Access for Patient Benefit (TAP-B, part of the UCL-RFH Biobank) under research ethics approval: 16/WA/0289 (Wales Research Ethics Committee 4). One case and one control were included on each slide. The Human Universal Cell Characterization core panel (960 genes) was used, supplemented with 8 additional genes to improve identification of cells of interest: CD1D , EREG , ETS2 , FCN1 , G0S2 , LYVE1 , MAP2K1 , MT1G . Segmentation was performed using the CosMx Human Universal Cell Segmentation Kit (RNA), Human IO PanCK/CD45 Kit (RNA) and Human CD68 Marker, Ch5 (RNA). Fields of view (FOVs) were tiled across all available regions (221 control, 378 PSC) and cyclic fluorescence in situ hybridization was performed using the CosMx SMI (Nanostring) system. Data were preprocessed on the AtoMx Spatial Informatics Platform, with images segmented to obtain cell boundaries, transcripts assigned to single cells, and a transcript by cell count matrix was obtained 97 . Expression matrices, transcript coordinates, polygon coordinates, FOV coordinates and cell metadata were exported, and quality control, normalization and cell-typing were performed using InSituType 98 —an R package developed to extract all the information available in every cell’s expression profile. A semi-supervised strategy was used to phenotype cells, incorporating the Liver Human Cell Atlas reference matrix. Spatial analysis of macrophage phenotypes was performed according to proximity from cholangiocytes (anchor cell type). Radius and nearest-neighbour analyses were performed using PhenoptR ( https://akoyabio.github.io/phenoptr/ ) with macrophage distribution from cholangiocytes binned in 100 µm increments up to 500 µm. Nearest-neighbour analysis was performed to determine the distance from cholangiocytes to the nearest inflammatory and non-inflammatory macrophage and vice versa.
To generate overlay images, raw transcript and image (morphology 2D) data were exported from AtoMx. Overlays of selected ETS2-target genes ( CXCL8 , S100A9 , CCL2 , CCL5 ) and fluorescent morphology markers were generated using napari (v.0.4.17, https://napari.org/stable/index.html ) on representative FOVs: FOV287 (PSC with involved duct), FOV294 (PSC background liver) and FOV55 (healthy liver).
Chr21q22 disease datasets
Publicly available raw RNA-seq data from the affected tissues of chr21q22-associated diseases (and controls from the same experiment) were downloaded from the GEO: IBD macrophages ( GSE123141 ), PSC liver ( GSE159676 ), ankylosing spondylitis synovium ( GSE41038 ). Reads were trimmed, filtered and aligned as described earlier. For each disease dataset, a ranked list of genes was obtained by differential expression analysis between cases and controls using limma with voom transformation. For IBD macrophages, only IBD samples with active disease were included. fGSEA using ETS2-regulated gene lists was performed as described.
LINCS signatures
A total of 31,027 lists of downregulated genes after exposure of a cell line to a small molecule was obtained from the NIH LINCS database 7 (downloaded in January 2021). These were used as gene sets for fGSEA (as described) with a ranked list of genes obtained by differential expression analysis between ETS2 -edited and unedited TPP macrophages (gRNA1) using limma with voom transformation and donor as a covariate. Drug classes for gene sets with FDR-adjusted P < 0.05 were manually assigned on the basis of known mechanisms of action.
MEK inhibition in TPP macrophages
TPP macrophages were generated as described previously. On day 4 of culture, PD-0325901 (0.5 μM, Sigma-Aldrich) or vehicle (DMSO) was added. Cells were collected on day 6 and RNA was extracted and sequenced as described.
Colonic biopsy explant culture
During colonoscopy, intestinal mucosal biopsies (6 per donor) were collected from ten patients with IBD (seven patients with ulcerative colitis, three patients with Crohn’s disease). All had endoscopically active disease and were not receiving immunosuppressive or biologic therapies. All biopsies were collected from a single inflamed site. All patients provided written informed consent. Ethical approval was provided by the London–Brent Regional Ethics Committee (21/LO/0682). Biopsies were collected into Opti-MEM and, within 1 h, were weighed and placed in pairs onto a Transwell insert (Thermo Fisher Scientific), designed to create an air–liquid interface 99 , in a 24-well plate. Each well contained 1 ml medium and was supplemented with either DMSO (vehicle control), PD-0325901 (0.5 μM) or infliximab (10 μg ml −1 ; MSD). Medium was as follows: Opti-MEM I (Gibco), GlutaMax (Thermo Fisher Scientific), 10% FBS (Thermo Fisher Scientific), MEM non-essential amino acids (Thermo Fisher Scientific), 1% sodium pyruvate (Thermo Fisher Scientific), 1% penicillin–streptomycin (Thermo Fisher Scientific) and 50 μg ml −1 gentamicin (Merck). After 18 h, the supernatants and biopsies were snap-frozen. The supernatant cytokine concentrations were quantified using the LEGENDplex Human Inflammation Panel (BioLegend). RNA was extracted from biopsies and libraries were prepared as described earlier ( n = 9, RNA from one donor was too degraded). Sequencing was performed on the NovaSeq 6000 system (100 bp paired-end reads). Data were processed as described earlier and GSVA was performed for ETS2-regulated genes and biopsy-derived signatures of IBD-associated inflammation 46 .
Chr21q22 genotypes in archaic humans
Using publicly available genomes from seven Neanderthal individuals 100 , 101 , 102 , 103 , one Denisovan individual 104 , and one Neanderthal and Denisovan F1 individual 105 , genotypes were called at the disease-associated chr21q22 candidate SNPs from the respective BAM files using bcftools mpileup with base and mapping quality options -q 20 -Q 20 -C 50 and using bcftools call -m -C alleles, specifying the two alleles expected at each site in a targets file (-T option). From the resulting .vcf file, the number of reads supporting the reference and alternative alleles was extracted and stored in the ‘DP4’ field.
Inference of Relate genealogy at rs2836882
Genome-wide genealogies, previously inferred for samples of the Simons Genome Diversity Project 106 using Relate 107 , 108 ( https://reichdata.hms.harvard.edu/pub/datasets/sgdp/ ), were downloaded from https://www.dropbox.com/sh/2gjyxe3kqzh932o/AAAQcipCHnySgEB873t9EQjNa?dl=0 . Using the inferred genealogies, the genealogy at rs2836882 (chr21:40466570) was plotted using the TreeView module of Relate.
Data presentation
The following R packages were used to create figures: GenomicRanges 109 , EnhancedVolcano 110 , ggplot2 (ref. 111 ), gplots 112 , karyoploteR 113 .
Statistical methodology
Statistical methods used in MPRA analysis, fGSEA and SNPsea are described above. For other analyses, comparison of continuous variables between two groups was performed using Wilcoxon matched-pairs tests (paired) or Mann–Whitney U -tests (unpaired) for nonparametric data or a t -tests for parametric data. Comparison against a hypothetical value was performed using Wilcoxon signed-rank tests for nonparametric data or one-sample t -tests for parametric data. A Shapiro–Wilk test was used to confirm normality. Two-sided tests were used as standard unless a specific hypothesis was being tested. Sample sizes are provided in the main text and figure captions.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The datasets produced in this study are accessible at the following repositories: MPRA (GEO: GSE229472 ), RNA-seq data of ETS2 or chr21q22-edited TPP macrophages (EGA: EGAD00001011338 ), RNA-seq data of ETS2 overexpression (EGA: EGAD00001011341 ), RNA-seq data of MEK-inhibitor-treated TPP macrophages (EGA: EGAD00001011337 ), H3K27ac ChIP–seq data in TPP macrophages (EGA: EGAD00001011351 ), ATAC–seq and H3K27ac ChIP–seq data in ETS2 -overexpressing or -edited macrophages (EGA: EGAD50000000154 ), ETS2 CUT&RUN data (EGA: EGAD00001011349 ), biopsy RNA-seq data (EGA: EGAD00001011333 ). MetaboLights: Metabolomics (MTBLS7665). The counts table for CosMx is provided at Zenodo ( https://zenodo.org/records/10707942 ) 114 . The phenotype and genotype data used for the PRS analysis are available on application to the IBD Bioresource ( https://www.ibdbioresource.nihr.ac.uk/ ). Source data are provided with this paper.
Code availability
Code to reproduce analyses are available at GitHub ( https://github.com/JamesLeeLab/chr21q22_manuscript ; https://github.com/chr1swallace/ibd-ets2-analysis ; https://github.com/qzhang314/PRS_IBD_subpheno ) 114 . Final code is deposited at Zenodo ( https://zenodo.org/records/10707942 ).
Miller, F. W. The increasing prevalence of autoimmunity and autoimmune diseases: an urgent call to action for improved understanding, diagnosis, treatment, and prevention. Curr. Opin. Immunol. 80 , 102266 (2023).
Article CAS PubMed Google Scholar
Dowden, H. & Munro, J. Trends in clinical success rates and therapeutic focus. Nat. Rev. Drug Discov. 18 , 495–496 (2019).
de Lange, K. M. et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat. Genet. 49 , 256–261 (2017).
Article PubMed PubMed Central Google Scholar
International Genetics of Ankylosing Spondylitis Consortium et al. Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat. Genet. 45 , 730–738 (2013).
Article Google Scholar
Ji, S. G. et al. Genome-wide association study of primary sclerosing cholangitis identifies new risk loci and quantifies the genetic relationship with inflammatory bowel disease. Nat. Genet. 49 , 269–273 (2017).
Ortiz-Fernandez, L. et al. Identification of susceptibility loci for Takayasu arteritis through a large multi-ancestral genome-wide association study. Am. J. Hum. Genet. 108 , 84–99 (2021).
Stathias, V. et al. LINCS Data Portal 2.0: next generation access point for perturbation-response signatures. Nucleic Acids Res. 48 , D431–D439 (2020).
Harrison, R. K. Phase II and phase III failures: 2013–2015. Nat. Rev. Drug Discov. 15 , 817–818 (2016).
Claussnitzer, M. et al. A brief history of human disease genetics. Nature 577 , 179–189 (2020).
Article CAS PubMed PubMed Central ADS Google Scholar
King, E. A., Davis, J. W. & Degner, J. F. Are drug targets with genetic support twice as likely to be approved? Revised estimates of the impact of genetic support for drug mechanisms on the probability of drug approval. PLoS Genet. 15 , e1008489 (2019).
Cader, M. Z. et al. C13orf31 (FAMIN) is a central regulator of immunometabolic function. Nat. Immunol. 17 , 1046–1056 (2016).
Article CAS PubMed PubMed Central Google Scholar
Murthy, A. et al. A Crohn’s disease variant in Atg16l1 enhances its degradation by caspase 3. Nature 506 , 456–462 (2014).
Article CAS PubMed ADS Google Scholar
Hnisz, D. et al. Super-enhancers in the control of cell identity and disease. Cell 155 , 934–947 (2013).
Park, M. D., Silvin, A., Ginhoux, F. & Merad, M. Macrophages in health and disease. Cell 185 , 4259–4279 (2022).
Kugathasan, S. et al. Loci on 20q13 and 21q22 are associated with pediatric-onset inflammatory bowel disease. Nat. Genet. 40 , 1211–1215 (2008).
Xue, J. et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity 40 , 274–288 (2014).
Kuo, D. et al. HBEGF + macrophages in rheumatoid arthritis induce fibroblast invasiveness. Sci. Transl. Med. 11 , eaau8587 (2019).
Melnikov, A. et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat. Biotechnol. 30 , 271–277 (2012).
Nerlov, C. & Graf, T. PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev. 12 , 2403–2412 (1998).
Minderjahn, J. et al. Mechanisms governing the pioneering and redistribution capabilities of the non-classical pioneer PU.1. Nat. Commun. 11 , 402 (2020).
Martinez, L. A. Mutant p53 and ETS2, a tale of reciprocity. Front. Oncol. 6 , 35 (2016).
Wei, G. et al. Activated Ets2 is required for persistent inflammatory responses in the motheaten viable model. J. Immunol. 173 , 1374–1379 (2004).
Zhao, J., Huang, K., Peng, H. Z. & Feng, J. F. Protein C-ets-2 epigenetically suppresses TLRs-induced interleukin 6 production in macrophages. Biochem. Biophys. Res. Commun. 522 , 960–964 (2020).
Chung, S. W., Chen, Y. H. & Perrella, M. A. Role of Ets-2 in the regulation of heme oxygenase-1 by endotoxin. J. Biol. Chem. 280 , 4578–4584 (2005).
Quinn, S. R. et al. The role of Ets2 transcription factor in the induction of microRNA-155 (miR-155) by lipopolysaccharide and its targeting by interleukin-10. J. Biol. Chem. 289 , 4316–4325 (2014).
Ma, X. et al. Ets2 suppresses inflammatory cytokines through MAPK/NF-κB signaling and directly binds to the IL-6 promoter in macrophages. Aging 11 , 10610–10625 (2019).
Aperlo, C., Pognonec, P., Stanley, E. R. & Boulukos, K. E. Constitutive c-ets2 expression in M1D + myeloblast leukemic cells induces their differentiation to macrophages. Mol. Cell. Biol. 16 , 6851–6858 (1996).
Henkel, G. W. et al. PU.1 but not ets-2 is essential for macrophage development from embryonic stem cells. Blood 88 , 2917–2926 (1996).
Mittal, M., Siddiqui, M. R., Tran, K., Reddy, S. P. & Malik, A. B. Reactive oxygen species in inflammation and tissue injury. Antioxid. Redox Signal. 20 , 1126–1167 (2014).
Kelly, B. & O’Neill, L. A. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. 25 , 771–784 (2015).
Martin, J. C. et al. Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell 178 , 1493–1508 (2019).
Peloquin, J. M. et al. Characterization of candidate genes in inflammatory bowel disease-associated risk loci. JCI Insight 1 , e87899 (2016).
Sazonovs, A. et al. Large-scale sequencing identifies multiple genes and rare variants associated with Crohn’s disease susceptibility. Nat. Genet. 54 , 1275–1283 (2022).
Slowikowski, K., Hu, X. & Raychaudhuri, S. SNPsea: an algorithm to identify cell types, tissues and pathways affected by risk loci. Bioinformatics 30 , 2496–2497 (2014).
Cramer, T. et al. HIF-1α is essential for myeloid cell-mediated inflammation. Cell 112 , 645–657 (2003).
Tannahill, G. M. et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 496 , 238–242 (2013).
Shiratori, R. et al. Glycolytic suppression dramatically changes the intracellular metabolic profile of multiple cancer cell lines in a mitochondrial metabolism-dependent manner. Sci. Rep. 9 , 18699 (2019).
Landt, S. G. et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 22 , 1813–1831 (2012).
Basuyaux, J. P., Ferreira, E., Stehelin, D. & Buttice, G. The Ets transcription factors interact with each other and with the c-Fos/c-Jun complex via distinct protein domains in a DNA-dependent and -independent manner. J. Biol. Chem. 272 , 26188–26195 (1997).
Sevilla, L. et al. Bcl-XL expression correlates with primary macrophage differentiation, activation of functional competence, and survival and results from synergistic transcriptional activation by Ets2 and PU.1. J. Biol. Chem. 276 , 17800–17807 (2001).
Kong, L. et al. The landscape of immune dysregulation in Crohn’s disease revealed through single-cell transcriptomic profiling in the ileum and colon. Immunity 56 , 444–458 (2023).
Chapuy, L. et al. Two distinct colonic CD14 + subsets characterized by single-cell RNA profiling in Crohn’s disease. Mucosal Immunol. 12 , 703–719 (2019).
Newman, J. A., Cooper, C. D., Aitkenhead, H. & Gileadi, O. Structural insights into the autoregulation and cooperativity of the human transcription factor Ets-2. J. Biol. Chem. 290 , 8539–8549 (2015).
Liu, H. et al. ERK differentially regulates Th17- and T reg -cell development and contributes to the pathogenesis of colitis. Eur. J. Immunol. 43 , 1716–1726 (2013).
Koboziev, I., Karlsson, F., Zhang, S. & Grisham, M. B. Pharmacological intervention studies using mouse models of the inflammatory bowel diseases: translating preclinical data into new drug therapies. Inflamm. Bowel Dis. 17 , 1229–1245 (2011).
Article PubMed Google Scholar
Argmann, C. et al. Biopsy and blood-based molecular biomarker of inflammation in IBD. Gut 72 , 1271–1287 (2023).
Malle, L. et al. Autoimmunity in Down’s syndrome via cytokines, CD4 T cells and CD11c + B cells. Nature 615 , 305–314 (2023).
Feagan, B. G. et al. Guselkumab plus golimumab combination therapy versus guselkumab or golimumab monotherapy in patients with ulcerative colitis (VEGA): a randomised, double-blind, controlled, phase 2, proof-of-concept trial. Lancet Gastroenterol. Hepatol. 8 , 307–320 (2023).
Friedrich, M. et al. IL-1-driven stromal-neutrophil interactions define a subset of patients with inflammatory bowel disease that does not respond to therapies. Nat. Med. 27 , 1970–1981 (2021).
Klesse, L. J. et al. The use of MEK inhibitors in neurofibromatosis type 1-associated tumors and management of toxicities. Oncologist 25 , e1109–e1116 (2020).
Zou, Y., Carbonetto, P., Wang, G. & Stephens, M. Fine-mapping from summary data with the “sum of single effects” model. PLoS Genet. 18 , e1010299 (2022).
1000 Genomes Consortium. A map of human genome variation from population-scale sequencing. Nature 467 , 1061–1073 (2010).
Kerimov, N. et al. A compendium of uniformly processed human gene expression and splicing quantitative trait loci. Nat. Genet. 53 , 1290–1299 (2021).
Fairfax, B. P. et al. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science 343 , 1246949 (2014).
Quach, H. et al. Genetic adaptation and Neandertal admixture shaped the immune system of human populations. Cell 167 , 643–656 (2016).
Chen, L. et al. Genetic drivers of epigenetic and transcriptional variation in human immune cells. Cell 167 , 1398–1414 (2016).
Nedelec, Y. et al. Genetic ancestry and natural selection drive population differences in immune responses to pathogens. Cell 167 , 657–669 (2016).
Alasoo, K. et al. Shared genetic effects on chromatin and gene expression indicate a role for enhancer priming in immune response. Nat. Genet. 50 , 424–431 (2018).
Wallace, C. A more accurate method for colocalisation analysis allowing for multiple causal variants. PLoS Genet. 17 , e1009440 (2021).
Bourges, C. et al. Resolving mechanisms of immune-mediated disease in primary CD4 T cells. EMBO Mol. Med. 12 , e12112 (2020).
Javierre, B. M. et al. Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell 167 , 1369–1384 (2016).
Hanzelmann, S., Castelo, R. & Guinney, J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 14 , 7 (2013).
Peters, J. E. et al. Insight into genotype-phenotype associations through eQTL mapping in multiple cell types in health and immune-mediated disease. PLoS Genet. 12 , e1005908 (2016).
Conant, D. et al. Inference of CRISPR edits from Sanger trace data. CRISPR J. 5 , 123–130 (2022).
Kalita, C. A. et al. QuASAR-MPRA: accurate allele-specific analysis for massively parallel reporter assays. Bioinformatics 34 , 787–794 (2018).
de Santiago, I. et al. BaalChIP: Bayesian analysis of allele-specific transcription factor binding in cancer genomes. Genome Biol. 18 , 39 (2017).
Corces, M. R. et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14 , 959–962 (2017).
Calderon, D. et al. Landscape of stimulation-responsive chromatin across diverse human immune cells. Nat. Genet. 51 , 1494–1505 (2019).
Reske, J. J., Wilson, M. R. & Chandler, R. L. ATAC-seq normalization method can significantly affect differential accessibility analysis and interpretation. Epigenet. Chromatin 13 , 22 (2020).
Article CAS Google Scholar
Brown, A. C. et al. Comprehensive epigenomic profiling reveals the extent of disease-specific chromatin states and informs target discovery in ankylosing spondylitis. Cell Genom. 3 , 100306 (2023).
Lienhard, M., Grimm, C., Morkel, M., Herwig, R. & Chavez, L. MEDIPS: genome-wide differential coverage analysis of sequencing data derived from DNA enrichment experiments. Bioinformatics 30 , 284–286 (2014).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 , 139–140 (2010).
Randzavola, L. O. et al. EROS is a selective chaperone regulating the phagocyte NADPH oxidase and purinergic signalling. eLife 11 , e76387 (2022).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37 , 907–915 (2019).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25 , 2078–2079 (2009).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30 , 923–930 (2014).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43 , e47 (2015).
Korotkevich, G. et al. Fast gene set enrichment analysis. Preprint at bioRxiv https://doi.org/10.1101/060012 (2021).
Qing, G. et al. Single-cell RNA sequencing revealed CD14 + monocytes increased in patients with Takayasu’s arteritis requiring surgical management. Front. Cell Dev. Biol. 9 , 761300 (2021).
Smillie, C. S. et al. Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell 178 , 714–730 (2019).
Gao, K. M. et al. Human nasal wash RNA-Seq reveals distinct cell-specific innate immune responses in influenza versus SARS-CoV-2. JCI Insight 6 , e152288 (2021).
Yang, Q. et al. The interaction of macrophages and CD8 T cells in bronchoalveolar lavage fluid is associated with latent tuberculosis infection. Emerg. Microbes Infect. 12 , 2239940 (2023).
Reyes, M. et al. An immune-cell signature of bacterial sepsis. Nat. Med. 26 , 333–340 (2020).
Mulder, K. et al. Cross-tissue single-cell landscape of human monocytes and macrophages in health and disease. Immunity 54 , 1883–1900 (2021).
Cassetta, L. et al. Human tumor-associated macrophage and monocyte transcriptional landscapes reveal cancer-specific reprogramming, biomarkers, and therapeutic targets. Cancer Cell 35 , 588–602 (2019).
Zernecke, A. et al. Integrated single-cell analysis-based classification of vascular mononuclear phagocytes in mouse and human atherosclerosis. Cardiovasc. Res. 119 , 1676–1689 (2023).
Ellinghaus, D. et al. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat. Genet. 48 , 510–518 (2016).
Renauer, P. A. et al. Identification of susceptibility loci in IL6, RPS9/LILRB3, and an intergenic locus on chromosome 21q22 in Takayasu arteritis in a genome-wide association study. Arthritis Rheumatol. 67 , 1361–1368 (2015).
Terao, C. et al. Genetic determinants and an epistasis of LILRA3 and HLA-B*52 in Takayasu arteritis. Proc. Natl Acad. Sci. USA 115 , 13045–13050 (2018).
Trubetskoy, V. et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature 604 , 502–508 (2022).
Bussi, C. et al. Lysosomal damage drives mitochondrial proteome remodelling and reprograms macrophage immunometabolism. Nat. Commun. 13 , 7338 (2022).
Behrends, V., Tredwell, G. D. & Bundy, J. G. A software complement to AMDIS for processing GC-MS metabolomic data. Anal. Biochem. 415 , 206–208 (2011).
Meers, M. P., Bryson, T. D., Henikoff, J. G. & Henikoff, S. Improved CUT&RUN chromatin profiling tools. eLife 8 , e46314 (2019).
Leporcq, C. et al. TFmotifView: a webserver for the visualization of transcription factor motifs in genomic regions. Nucleic Acids Res. 48 , W208–W217 (2020).
Weirauch, M. T. et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158 , 1431–1443 (2014).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184 , 3573–3587 (2021).
He, S. et al. High-plex imaging of RNA and proteins at subcellular resolution in fixed tissue by spatial molecular imaging. Nat. Biotechnol. 40 , 1794–1806 (2022).
Danaher, P. et al. InSituType: likelihood-based cell typing for single cell spatial transcriptomics. Preprint at bioRxiv https://doi.org/10.1101/2022.10.19.512902 (2022).
Vadstrup, K. et al. Validation and optimization of an ex vivo assay of intestinal mucosal biopsies in Crohn’s disease: reflects inflammation and drug effects. PLoS ONE 11 , e0155335 (2016).
Prufer, K. et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505 , 43–49 (2014).
Article PubMed ADS Google Scholar
Prufer, K. et al. A high-coverage Neandertal genome from Vindija Cave in Croatia. Science 358 , 655–658 (2017).
Article PubMed PubMed Central ADS Google Scholar
Hajdinjak, M. et al. Reconstructing the genetic history of late Neanderthals. Nature 555 , 652–656 (2018).
Mafessoni, F. et al. A high-coverage Neandertal genome from Chagyrskaya Cave. Proc. Natl Acad. Sci. USA 117 , 15132–15136 (2020).
Meyer, M. et al. A high-coverage genome sequence from an archaic Denisovan individual. Science 338 , 222–226 (2012).
Slon, V. et al. The genome of the offspring of a Neanderthal mother and a Denisovan father. Nature 561 , 113–116 (2018).
Mallick, S. et al. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature 538 , 201–206 (2016).
Speidel, L., Forest, M., Shi, S. & Myers, S. R. A method for genome-wide genealogy estimation for thousands of samples. Nat. Genet. 51 , 1321–1329 (2019).
Speidel, L. et al. Inferring population histories for ancient genomes using genome-wide genealogies. Mol. Biol. Evol. 38 , 3497–3511 (2021).
Lawrence, M. et al. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 9 , e1003118 (2013).
Blighe, K. et al. EnhancedVolcano: publication-ready volcano plots with enhanced colouring and labeling. Bioconductor https://doi.org/10.18129/B9.bioc.EnhancedVolcano (2023).
Hadley, W. Ggplot2 (Springer, 2016).
Warnes, G. et al. gplots: various R programming tools for plotting data. CRAN https://CRAN.R-project.org/package=gplots (2022).
Gel, B. & Serra, E. karyoploteR: an R/Bioconductor package to plot customizable genomes displaying arbitrary data. Bioinformatics 33 , 3088–3090 (2017).
Stankey, C. T. et al. Data for ‘A disease-associated gene desert directs macrophage inflammation through ETS2’. Zenodo https://doi.org/10.5281/zenodo.10707942 (2024).
Download references
Acknowledgements
We thank the members of the Lee laboratory, K. Slowikowski and A. Kaser for discussions; G. Stockinger, C. Vinuesa, C. Swanton, R. Patani and C. Reis e Sousa for reading the manuscript; C. Cheshire and the staff at the Francis Crick Institute Advanced Sequencing Facility and Flow Cytometry STP for technical support; L. Lucaciu for help with patient recruitment; RFH PITU nurses for assistance obtaining infliximab; the members of Tissue Access for Patient Benefit (TAP-B) for providing liver samples; NIHR BioResource volunteers for their participation; and the NIHR BioResource centres, NHS Blood and Transplant, and NHS staff for their contributions. This work was supported by Crohn’s and Colitis UK (M2018-3), the Wellcome Trust (Sir Henry Wellcome Fellowship to L.S., 220457/Z/20/Z; Investigator Award to P.S., 217223/Z/19/Z; Senior Fellowship to C.W., WT220788; Clinical Research Career Development Fellowship to M.Z.C., 222056/Z/20/Z; Wellcome-Beit Prize Clinical Career Development Fellowship to D.C.T., 206617/A/17/A; and Intermediate Clinical Fellowship to J.C.L., 105920/Z/14/Z), and the Francis Crick Institute, which receives its core funding from Cancer Research UK (CC2219, FC001595), the UK Medical Research Council (CC2219, FC001595) and the Wellcome Trust (CC2219, FC001595). L.M.H. is supported by the Charité–Universitätsmedizin Berlin and the Berlin Institute of Health Charité (Clinician-Scientist Program); A.J.C. by the Medical Research Council (MR/V029711/1); A.L. by a Lord Kelvin/Adam Smith Leadership Grant; A.H.S. by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIH, R01:AR070148); N.B.J. by Cancer Research UK (C55370/A25813); T.Z. by the Chinese Scholarship Council (202308060128); A.Q. by the NIHR UCLH/UCL BRC; J.C.K. by Versus Arthritis (program grant, 20773), Janssen Oxford Translational fellowships and NIHR Oxford BRC; P.S. by the European Molecular Biology Organisation, the Vallee Foundation and the European Research Council (852558); C.W. by the Medical Research Council (MC UU 00002/4), GSK, MSD and the NIHR Cambridge BRC (BRC-1215-20014); and D.C.T. by the Sidharth Burman endowment. J.C.L. is a Lister Institute Prize Fellow. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. Experimental schematics in Figs. 1d , 2a and 3a and Extended Data Figs. 3a , 4a,b,e and 7g,h were created using BioRender. For the purpose of open access, the authors have applied a CC BY public copyright licence to any author accepted manuscript version arising from this submission.
Open Access funding provided by The Francis Crick Institute.
Author information
These authors contributed equally: C. T. Stankey, C. Bourges, L. M. Haag
Authors and Affiliations
Genetic Mechanisms of Disease Laboratory, The Francis Crick Institute, London, UK
C. T. Stankey, C. Bourges, T. Turner-Stokes, A. P. Piedade, I. Papa, E. C. Parkes & J. C. Lee
Department of Immunology and Inflammation, Imperial College London, London, UK
C. T. Stankey, T. Turner-Stokes & L. O. Randzavola
Washington University School of Medicine, St Louis, MO, USA
C. T. Stankey
Division of Gastroenterology, Infectious Diseases and Rheumatology, Charité–Universitätsmedizin Berlin, Berlin, Germany
Department of Gastroenterology, Royal Free Hospital, London, UK
C. Palmer-Jones, A. P. Rochford, C. D. Murray & J. C. Lee
Institute for Liver and Digestive Health, Division of Medicine, University College London, London, UK
C. Palmer-Jones, F. Saffioti, D. Thorburn, A. P. Rochford, C. D. Murray & J. C. Lee
Metabolomics STP, The Francis Crick Institute, London, UK
M. Silva dos Santos & J. I. MacRae
Genomics of Inflammation and Immunity Group, Human Genetics Programme, Wellcome Sanger Institute, Hinxton, UK
Wolfson Wohl Cancer Centre, School of Cancer Sciences, University of Glasgow, Glasgow, UK
A. J. Cameron, A. Legrini, T. Zhang, C. S. Wood & N. B. Jamieson
NanoString Technologies, Seattle, WA, USA
F. N. New & P. Divakar
Ancient Genomics Laboratory, The Francis Crick Institute, London, UK
L. Speidel & P. Skoglund
Genetics Institute, University College London, London, UK
Wellcome Centre for Human Genetics, University of Oxford, Oxford, UK
A. C. Brown & J. C. Knight
The Sheila Sherlock Liver Centre, Royal Free Hospital, London, UK
A. Hall, F. Saffioti & D. Thorburn
Department of Cellular Pathology, Royal Free Hospital, London, UK
A. Hall & A. Quaglia
Cambridge Institute of Therapeutic Immunology and Infectious Disease, University of Cambridge, Cambridge, UK
W. Edwards, M. Z. Cader, C. Wallace & D. C. Thomas
Department of Internal Medicine, Division of Rheumatology, Marmara University, Istanbul, Turkey
H. Direskeneli
Systemic Autoimmunity Branch, NIAMS, National Institutes of Health, Bethesda, MD, USA
P. C. Grayson
Department of Rheumatology, Zhongshan Hospital, Fudan University, Shanghai, China
Division of Rheumatology, Department of Medicine, University of Pennsylvania, Philadelphia, PA, USA
P. A. Merkel
Division of Epidemiology, Department of Biostatistics, Epidemiology and Informatics, University of Pennsylvania, Philadelphia, PA, USA
Department of Physiology, Istanbul University, Istanbul Faculty of Medicine, Istanbul, Turkey
G. Saruhan-Direskeneli
Division of Rheumatology, Department of Pediatrics, University of Pittsburgh, Pittsburgh, PA, USA
A. H. Sawalha
Division of Rheumatology and Clinical Immunology, Department of Medicine, University of Pittsburgh, Pittsburgh, PA, USA
Lupus Center of Excellence, University of Pittsburgh, Pittsburgh, PA, USA
Department of Immunology, University of Pittsburgh, Pittsburgh, PA, USA
Department of Biomedical and Clinical Sciences, Milan University, Milan, Italy
E. Tombetti
Internal Medicine and Rheumatology, ASST FBF-Sacco, Milan, Italy
UCL Cancer Institute, London, UK
Chinese Academy of Medical Sciences Institute, Nuffield Department of Medicine, University of Oxford, Oxford, UK
J. C. Knight
NIHR Comprehensive Biomedical Research Centre, Oxford, UK
Experimental Histopathology STP, The Francis Crick Institute, London, UK
M. Green & E. Nye
Department of Medicine, University of Cambridge, Cambridge, UK
M. Z. Cader & D. C. Thomas
MRC Biostatistics Unit, Cambridge Institute of Public Health, Cambridge, UK
You can also search for this author in PubMed Google Scholar
Contributions
Conceptualization: J.I.M., N.B.J., P.S., M.Z.C., C.W., D.C.T. and J.C.L. Methodology: C.T.S., C.B., M.S.d.S., M.G., E.N., J.I.M., C.W. and J.C.L. Software: C.B., M.S.d.S., Q.Z., A.J.C., A.L., T.Z., C.S.W., L.S., J.I.M., N.B.J., P.S., C.W. and J.C.L. Investigation: C.T.S., C.B., L.M.H., T.T.-S., A.P.P., I.P., M.S.d.S., L.O.R., A.C.B., E.C.P., W.E., M.G., C.D.M. and J.C.L. Resources: C.T.S., C.B., C.P.-J., A.H., F.S., A.Q., D.T., A.P.R., C.D.M. and J.C.L. Formal analysis: C.T.S., C.B., M.S.d.S., Q.Z., A.J.C., A.L., T.Z., F.N.N., L.S., P.D., C.W. and J.C.L. Writing—original draft: C.T.S., C.B. and J.C.L. Writing—review and editing: all of the authors. Funding acquisition: J.C.L. Supervision: J.C.K., J.I.M., N.B.J., P.S., C.W., D.C.T. and J.C.L.
Corresponding author
Correspondence to J. C. Lee .
Ethics declarations
Competing interests.
C.T.S., C.B. and J.C.L. are listed as co-inventors on a patent application related to this work. C.W. holds a part-time position at GSK. GSK had no role in the design or conduct of this study. F.N.N. and P.D. are employees and shareholders of NanoString Technologies. NanoString had no role in the design or conduct of this study. The other authors declare no competing interests.
Peer review
Peer review information.
Nature thanks Joachim Schultze and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended data fig. 1 colocalisation between genetic associations at chr21q22..
a . Example comparison of genetic associations at chr21q22: IBD and ETS2 eQTL in unstimulated monocytes. Plot adapted from locuscomparer. b . Tukey box-and-whisker plot depicting ETS2 expression stratified by rs2836882 genotype in unstimulated monocytes (AA, n = 39; AG, n = 142; GG, n = 233) 54 . P -value is as reported in index study. c . Radar plot of representative colocalization results for the indicated genetic associations compared to IBD. Posterior probability of independent causal variants, PP.H3, dark blue; posterior probability of shared causal variant, PP.H4, light blue. PP.H4 > 0.5 was used to call colocalisation (denoted by dashed line). Labels are coloured according to class of data (indicated in the key). Asterisks denote colocalisation. Data sources are: IBD 3 , PSC 5 , AS 4 , Takayasu Arteritis 6 , BLUEPRINT 56 , Fairfax 54 , Quach 55 , Nedelec 57 , Alasoo 58 .
Extended Data Fig. 2 CRISPR-Cas9 editing of the chr21q22 locus and ETS2 in monocytes.
a . Cas9 gRNAs were designed to flank the chr21q22 enhancer region at the indicated sites. b . Representative bioanalyzer trace of PCR-amplified target region following monocyte CRISPR/Cas9 editing with an equimolar mix of RNPs containing 5′ and 3′ chr21q22 gRNAs. Example editing efficiency calculation shown. c . Editing efficiency at the chr21q22 locus. Mean enhancer deletion: 42.4% (n = 11). d . Location and sequence of gRNAs used to disrupt ETS2 . e . ETS2 editing efficiency. gRNA1 (mean), 89.7% (n = 31); gRNA2 (mean), 78.6% (n = 14). f . ETS2 expression (relative to NTC) following CRISPR/Cas9 editing, measured by qPCR (housekeeping gene PPIA ; equivalent results with other housekeeping genes; n = 10). g . Viability following monocyte nucleofection with Cas9 RNPs and macrophage differentiation. Mean values: NTC, 97.9%; gRNA1: 98.3%; gRNA2, 98.6% (n = 6). h . Expression of myeloid lineage markers following ETS2 editing and TPP differentiation (n = 5). Gating strategy shown in Supplementary Information Fig. 2 . i . GSVA enrichment scores for 67 different monocyte/macrophage activation conditions to identify stimuli that phenocopy CD14+ monocytes/macrophages from IBD patients. j . Chromatin accessibility in ETS2-edited versus unedited inflammatory macrophages (n = 3). k . Enhancer activity (H3K27ac) in ETS2-edited versus unedited inflammatory macrophages (n = 3). P values calculated using edgeR (two-sided) in j , k . Red points denote adjusted P -value (P adj ) < 0.1, grey points NS. Error bars are mean±SEM in c , e - h . * P < 0.05. NTC: non-targeting control.
Extended Data Fig. 3 Optimization of MPRA and mRNA overexpression in primary human macrophages.
a . Schematic of MPRA. A library of oligonucleotides (each containing a genomic sequence and unique barcode, separated by restriction enzyme sites) is cloned into a pGL4.10 M cloning vector. A promoter and reporter gene are inserted using directional cloning. The resulting plasmids are transfected into primary human macrophages (TPP) and RNA is extracted after 24 h. Barcode abundance in cellular mRNA and input DNA library are quantified by high-throughput sequencing, and mRNA barcode counts are normalized to corresponding counts in DNA library to assess expression-modulating activity. b . Identification of suitable promoters for MPRA in TPP macrophages. TPP macrophages were transfected with reporter vectors, each with GFP expression under the control of a different promoter. GFP expression was quantified by flow cytometry after 24 h. c . Adapted MPRA vector for use in primary human macrophages, containing RSV promoter. d . Heatmap showing pairwise correlation of expression-modulating activity of all constructs between donors. e . Principal component analysis of element counts (sum of barcodes tagging same genomic sequence) in mRNA from TPP macrophages (n = 8 donors; red) and four replicates of DNA vector (black). f . Primary human macrophages (M0) were transfected with different quantities of GFP mRNA using Lipofectamine MessengerMAX. GFP expression was quantified by flow cytometry 18 h after transfection. g . Cytokine secretion following ETS2 overexpression. Plot shows relative cytokine concentrations in macrophage supernatants ( ETS2 relative to control) following transfection with 500 ng mRNA (n = 11). Error bars are mean±SEM. One-sample t -test (two-tailed) * P < 0.05, ** P < 0.01. The diagram in a was created using BioRender.
Extended Data Fig. 4 Molecular effects of allelic variation at rs2836882 .
a . Schematic of PU.1 ChIP-genotyping assay to assess allele-specific PU.1 binding at rs2836882 in human macrophages. b . Schematic of standard curve generation by TaqMan genotyping various pre-defined ratios of risk and non-risk containing DNA sequences. c . Standard curve generated using different allelic ratios of 200-nt DNA geneblocks centred on either the major (risk) or minor (non-risk) rs2836882 allele. d . Allele-specific PU.1 binding at rs2836882 in TPP macrophages (one-sample t -test, two-sided, n = 5). Error bars represent mean±95%CI. e . Schematic of PU.1 MPRA-ChIP assay to assess allele-specific PU.1 binding at individual SNPs within chr21q22 enhancer. f . Allele-specific PU.1 binding at SNPs within chr21q22 enhancer in TPP macrophages. Data represents the allelic ratio of normalized PU.1 binding for constructs centred on the SNP allele from the MPRA library (fixed-effects meta-analysis of QuASAR-MPRA results, two-sided, n = 6). Box represents median (IQR), whiskers represent minima and maxima. g . Allele-specific ATAC-seq reads at rs2836882 in two deeply sequenced heterozygous TPP macrophage datasets (left: 154.7 million non-duplicate paired-end reads, right: 165.4 million non-duplicate paired-end reads). h . H3K27ac ChIP-seq data from risk (red) or non-risk (blue) allele homozygotes at rs2836882 (n = 4). i . Rank Ordering of Super-Enhancers (ROSE) analysis of H3K27ac ChIP-seq data from TPP macrophages from major (left) and minor (right) allele homozygotes. Dashed line denotes inflection point of curve, with enhancers above this point being denoted as super-enhancers. Red points indicate rs2836882 -containing chr21q22 enhancer. SE, super-enhancer. The diagrams in a , b and e were created using BioRender.
Extended Data Fig. 5 Functional effects of the chr21q22 enhancer.
a . Extracellular ROS production by unedited (NTC), chr21q22-edited, and ETS2 g1-edited TPP macrophages, quantified by chemiluminescence. Points represent relative area under curve for edited versus unedited cells (Wilcoxon signed-rank test, two-sided; n = 6). b . Cytokine secretion from inflammatory macrophages following deletion of the chr21q22 enhancer. Heatmap shows relative cytokine concentrations in the supernatants of chr21q22-edited TPP macrophages versus unedited (NTC) cells (Wilcoxon signed rank test, one-sided; n = 7). c . Representative flow cytometry histograms demonstrating phagocytosis of fluorescently-labelled zymosan particles by chr21q22-edited and unedited (NTC) TPP macrophages. d . Phagocytosis index for unedited and chr21q22-edited TPP macrophages, calculated as proportion of positive cells multiplied by mean fluorescence intensity of positive cells. Plot shows relative phagocytosis index for chr21q22-edited cells versus unedited cells (Wilcoxon signed-rank test two-sided; n = 7). e . Enrichment of differentially-expressed genes following deletion of the disease-associated chr21q22 locus (upregulated genes, top; downregulated genes, bottom) in ETS2 -edited versus unedited macrophages. P adj , FDR-adjusted P -value (two-sided). f . Tukey box-and-whisker plot depicting quantitative PCR of selected ETS2-target genes in resting (M0) macrophages from minor and major allele homozygote IBD patients (n = 22, expression normalized to PPIA and scaled to minimum 0, maximum 1). Mann-Whitney test (one-sided). * P < 0.05, ** P < 0.01, *** P < 0.001.
Extended Data Fig. 6 Polygenic Risk Score of 22 ETS2-regulated IBD-associated genes.
a . Summary of IBD BioResource cohorts used for PRS analysis. b . Association between PRS and age at diagnosis. c . Association between PRS and extent of ulcerative colitis (E1, proctitis; E2, left-sided; E3, extensive colitis). d . Association between PRS and Crohn’s disease location (L1, ileal; L2, colonic; L3, ileocolonic). L2 is associated with a milder disease phenotype. e . Association between PRS and perianal involvement in Crohn’s disease. f . Association between PRS and Crohn’s disease behaviour (B1, inflammatory; B2, stricturing; B3, fistulating). B2 and B3 represent more aggressive, complicated forms of Crohn’s disease. g . Association between PRS and response to anti-TNFα in Crohn’s disease and ulcerative colitis (PR, primary responder; PNR, primary non-responder). h . Association between PRS and need for surgery in Crohn’s disease and ulcerative colitis. Overall, higher PRS was associated with: earlier age at diagnosis, ileal or ileocolonic forms of Crohn’s disease, B2/B3 Crohn’s disease behaviour, and increased need for surgery in IBD. Analysis in b performed using linear regression. Analyses in c - h performed using logistic regression (with diagnosis as covariate in g and h ). SNPs included in PRS are listed in Extended Data Table 1 . i . Plot of enrichment statistic (standardized effect size) against statistical significance from SNPsea analysis of genes tagged by 241 IBD SNPs within ETS2 -regulated genes (red) and known IBD pathways (black). j . SNPsea analyses of SNPs associated with PSC, ankylosing spondylitis, Takayasu’s arteritis or Schizophrenia (negative control) within lists of ETS2-regulated genes–either upregulated by ETS2 overexpression, downregulated by ETS2 disruption, or downregulated following chr21q22 deletion (all FDR < 0.05). Dashed line denotes P < 0.05.
Extended Data Fig. 7 Effects of modulating ETS2.
a and b . Changes in total metabolite abundance ( a ) and percentage of label incorporation from 13 C-glucose ( b ) following ETS2 editing in TPP macrophages (n = 6). Colour depicts median log2 fold-change in ETS2 -edited macrophages relative to unedited macrophages (transfected with non-targeting control RNPs; NTC). Bold black border indicates P < 0.05 (Wilcoxon signed rank test, two-sided). c . Heatmap summarizing metabolic changes following ETS2 disruption. Colour depicts median log2 fold-change in ETS2 g1-edited cells relative to unedited cells (Wilcoxon signed rank test, two-sided, * P < 0.05). d . Phagocytosis index in unedited (NTC) and ETS2 -edited TPP macrophages treated with roxadustat (ROX) or vehicle. Phagocytosis index is calculated as proportion of positive cells multiplied by mean fluorescence intensity of positive cells (488 nm channel). Data normalized to phagocytosis index in unedited cells (n = 5). e . Extracellular ROS production by unedited (NTC) and ETS2 -edited TPP macrophages treated with ROX or vehicle – quantified using a chemiluminescence assay. Data represent log2 fold-change of area under curve (AUC) normalized to unedited (NTC) TPP macrophages (n = 5). f . TFmotifView enrichment results for motifs of transcription factors expressed in TPP macrophages (CPM > 0.5) within ETS2 CUT&RUN peaks. Results shown for all significantly enriched transcription factors (Bonferroni P value < 0.05, two-sided) with motifs in more than 10% peaks. g . Schematic of experiment to assess how ETS2 disruption affects the activity of the chr21q22 ETS2 enhancer in inflammatory (TPP) macrophages. h . Schematic of experiment to assess how ETS2 overexpression affects the activity of the chr21q22 ETS2 enhancer in resting (M0) macrophages. i . Normalized H3K27ac ChIP-seq read counts (edgeR fitted values) from chr21:40,465,000-40,470,000 in experiments depicted in g (left) and h (right) (edgeR P values, two-sided, n = 3 for each). Error bars in d and e represent mean±SEM. The diagrams in g and h were created using BioRender.
Extended Data Fig. 8 The transcriptional signature of ETS2 is detectable in affected tissues from chr21q22-linked diseases.
a . ETS2 expression in scRNA-seq clusters of myeloid cells from Crohn’s disease and healthy controls (upper panel). Relative contributions of single cells from Crohn’s disease or healthy controls to individual clusters (same UMAP dimensions as for combined analysis). b . Overlay of CosMx morphology 2D image data and raw transcripts of selected ETS2 target genes. Fluorescent morphology markers alone (top row), CXCL8 (cyan) and S1009A (yellow) transcripts (middle row), CCL5 (cyan) and CCL2 (yellow) transcripts (bottom row). Columns are representative examples of PSC with diseased ducts (left), PSC with uninflamed background liver (centre), and healthy liver (right). Size marker (white) on every field of view (FOV) denotes 50 µm. c . Gene set enrichment analysis (fGSEA) of genes downregulated following chr21q22 enhancer deletion or ETS2 disruption (gRNA1 or gRNA2) within intestinal macrophages from patients with active IBD (compared to control intestinal macrophages, n = 20; left), ankylosing spondylitis synovium (compared to control synovium, n = 15; centre), and PSC liver biopsies (compared to control liver biopsies, n = 17; right). P adj , FDR-adjusted P -value (two-sided).
Extended Data Fig. 9 Effect of MEK1/2 inhibition on ETS2- regulated genes.
a - c . Gene set enrichment analysis (fGSEA) in MEK1/2 inhibitor-treated TPP macrophages showing enrichment of gene sets upregulated (upper panel) or downregulated (lower panel) following ETS2 or chr21q22 editing (MEK1/2 inhibited using PD-0325901, 0.5 µM). Gene sets obtained from differential gene expression analysis (limma using voom transformation) following ETS2 disruption with gRNA1 ( a ), gRNA2 ( b ), or following chr21q22 deletion ( c ). d . fGSEA in intestinal biopsies from IBD patients showing enrichment of gene sets downregulated following ETS2 or chr21q22 editing in MEK inhibitor-treated biopsies. Upregulated gene sets were not enriched. e . Proportion and pathway analysis of MEK inhibitor-induced differentially expressed genes that have no evidence for being ETS2 targets in macrophages (incorporating differential expression from knockout or overexpression experiments and promoter / regulatory element binding from ETS2 CUT&RUN). P adj , FDR-adjusted P -value (two-sided).
Extended Data Fig. 10 Geographic distribution and history of rs2836882 .
a . rs2836882 allele frequency in modern global populations (data from 1000 Genomes Project, plotted using Geography of Genetic Variants browser: https://popgen.uchicago.edu/ggv/ ). b . Genotypes of candidate SNPs at chr21q22 (99% credible set) in archaic humans (Neanderthals and Denisovans). Colour depicts the proportion of reads containing ALT alleles, with a value close to 0 consistent with a homozygous REF (risk) genotype, a value close to 1 consistent with a homozygous ALT (non-risk) genotype, and an intermediate value indicating a potential heterozygous genotype. Number in each cell indicates the number of reads at that SNP in the indicated sample. Putative causal variant highlighted in red. c . Inferred genealogy of the age of the rs2836882 polymorphism – analysed using Relate. The diagram in a was created using the Geography of Genetic Variants browser.
Supplementary information
Supplementary figures.
Supplementary Fig. 1: uncropped Western blots from Fig. 2d. Two lanes were run for each sample: one lane to blot for vinculin and the NADPH oxidase components gp91phox, gp65 and p22phox, and one lane to blot for vinculin and the chaperone protein EROS. After transfer, the membranes were cut to blot for individual targets. Supplementary Fig. 2: example gating strategy. Example gating strategy for MPRA and macrophage phenotyping. Macrophages were gated by FSC-A/SSC-A and singlets were gated by FSC-A/FSC-H. Live cells were gated (and viability was quantified) using Live/Dead Fixable Aqua Dead Cell Stain.
Reporting Summary
Supplementary tables.
Supplementary Table 1: differentially expressed genes in primary macrophages after ETS2 or chr21q22 CRISPR–Cas9 editing. Supplementary Table 2: differentially expressed genes in primary macrophages after ETS2 overexpression. Supplementary Table 3: the primers and gRNA sequences used in this study.
Peer Review File
Source data, source data fig. 1, source data fig. 2, source data fig. 3, source data fig. 4, source data fig. 5, source data extended data fig. 1, source data extended data fig. 2, source data extended data fig. 3, source data extended data fig. 4, source data extended data fig. 5, source data extended data fig. 6, source data extended data fig. 7, source data extended data fig. 8, source data extended data fig. 9, source data extended data fig. 10, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Stankey, C.T., Bourges, C., Haag, L.M. et al. A disease-associated gene desert directs macrophage inflammation through ETS2. Nature 630 , 447–456 (2024). https://doi.org/10.1038/s41586-024-07501-1
Download citation
Received : 17 April 2023
Accepted : 01 May 2024
Published : 05 June 2024
Issue Date : 13 June 2024
DOI : https://doi.org/10.1038/s41586-024-07501-1
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
By submitting a comment you agree to abide by our Terms and Community Guidelines . If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing: Translational Research newsletter — top stories in biotechnology, drug discovery and pharma.
Have a language expert improve your writing
Run a free plagiarism check in 10 minutes, generate accurate citations for free.
- Knowledge Base
Methodology
- What is Secondary Research? | Definition, Types, & Examples
What is Secondary Research? | Definition, Types, & Examples
Published on January 20, 2023 by Tegan George . Revised on January 12, 2024.
Secondary research is a research method that uses data that was collected by someone else. In other words, whenever you conduct research using data that already exists, you are conducting secondary research. On the other hand, any type of research that you undertake yourself is called primary research .
Secondary research can be qualitative or quantitative in nature. It often uses data gathered from published peer-reviewed papers, meta-analyses, or government or private sector databases and datasets.
Table of contents
When to use secondary research, types of secondary research, examples of secondary research, advantages and disadvantages of secondary research, other interesting articles, frequently asked questions.
Secondary research is a very common research method, used in lieu of collecting your own primary data. It is often used in research designs or as a way to start your research process if you plan to conduct primary research later on.
Since it is often inexpensive or free to access, secondary research is a low-stakes way to determine if further primary research is needed, as gaps in secondary research are a strong indication that primary research is necessary. For this reason, while secondary research can theoretically be exploratory or explanatory in nature, it is usually explanatory: aiming to explain the causes and consequences of a well-defined problem.
Prevent plagiarism. Run a free check.
Secondary research can take many forms, but the most common types are:
Statistical analysis
Literature reviews, case studies, content analysis.
There is ample data available online from a variety of sources, often in the form of datasets. These datasets are often open-source or downloadable at a low cost, and are ideal for conducting statistical analyses such as hypothesis testing or regression analysis .
Credible sources for existing data include:
- The government
- Government agencies
- Non-governmental organizations
- Educational institutions
- Businesses or consultancies
- Libraries or archives
- Newspapers, academic journals, or magazines
A literature review is a survey of preexisting scholarly sources on your topic. It provides an overview of current knowledge, allowing you to identify relevant themes, debates, and gaps in the research you analyze. You can later apply these to your own work, or use them as a jumping-off point to conduct primary research of your own.
Structured much like a regular academic paper (with a clear introduction, body, and conclusion), a literature review is a great way to evaluate the current state of research and demonstrate your knowledge of the scholarly debates around your topic.
A case study is a detailed study of a specific subject. It is usually qualitative in nature and can focus on a person, group, place, event, organization, or phenomenon. A case study is a great way to utilize existing research to gain concrete, contextual, and in-depth knowledge about your real-world subject.
You can choose to focus on just one complex case, exploring a single subject in great detail, or examine multiple cases if you’d prefer to compare different aspects of your topic. Preexisting interviews , observational studies , or other sources of primary data make for great case studies.
Content analysis is a research method that studies patterns in recorded communication by utilizing existing texts. It can be either quantitative or qualitative in nature, depending on whether you choose to analyze countable or measurable patterns, or more interpretive ones. Content analysis is popular in communication studies, but it is also widely used in historical analysis, anthropology, and psychology to make more semantic qualitative inferences.
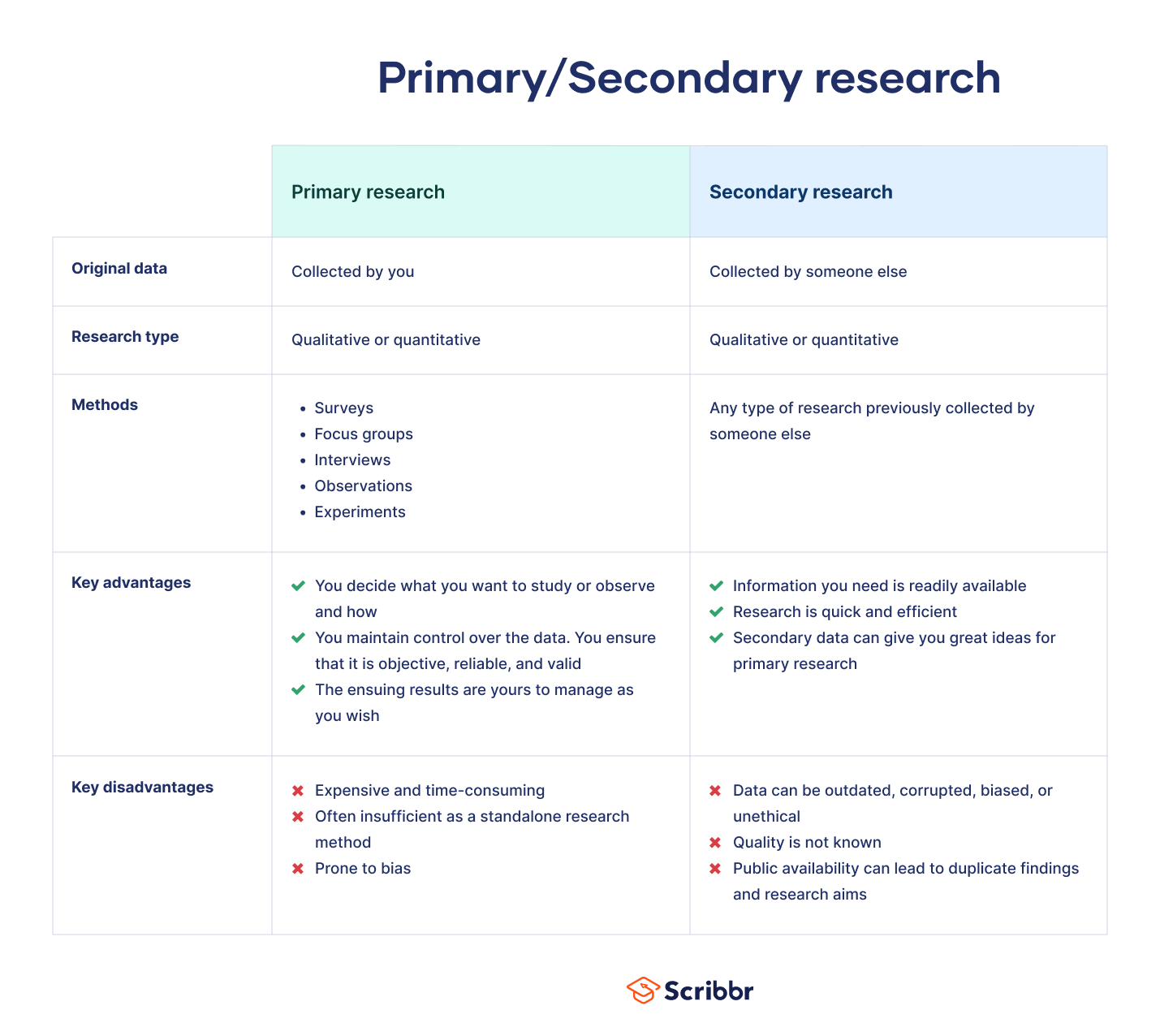
Secondary research is a broad research approach that can be pursued any way you’d like. Here are a few examples of different ways you can use secondary research to explore your research topic .
Secondary research is a very common research approach, but has distinct advantages and disadvantages.
Advantages of secondary research
Advantages include:
- Secondary data is very easy to source and readily available .
- It is also often free or accessible through your educational institution’s library or network, making it much cheaper to conduct than primary research .
- As you are relying on research that already exists, conducting secondary research is much less time consuming than primary research. Since your timeline is so much shorter, your research can be ready to publish sooner.
- Using data from others allows you to show reproducibility and replicability , bolstering prior research and situating your own work within your field.
Disadvantages of secondary research
Disadvantages include:
- Ease of access does not signify credibility . It’s important to be aware that secondary research is not always reliable , and can often be out of date. It’s critical to analyze any data you’re thinking of using prior to getting started, using a method like the CRAAP test .
- Secondary research often relies on primary research already conducted. If this original research is biased in any way, those research biases could creep into the secondary results.
Many researchers using the same secondary research to form similar conclusions can also take away from the uniqueness and reliability of your research. Many datasets become “kitchen-sink” models, where too many variables are added in an attempt to draw increasingly niche conclusions from overused data . Data cleansing may be necessary to test the quality of the research.
Here's why students love Scribbr's proofreading services
Discover proofreading & editing
If you want to know more about statistics , methodology , or research bias , make sure to check out some of our other articles with explanations and examples.
- Normal distribution
- Degrees of freedom
- Null hypothesis
- Discourse analysis
- Control groups
- Mixed methods research
- Non-probability sampling
- Quantitative research
- Inclusion and exclusion criteria
Research bias
- Rosenthal effect
- Implicit bias
- Cognitive bias
- Selection bias
- Negativity bias
- Status quo bias
A systematic review is secondary research because it uses existing research. You don’t collect new data yourself.
The research methods you use depend on the type of data you need to answer your research question .
- If you want to measure something or test a hypothesis , use quantitative methods . If you want to explore ideas, thoughts and meanings, use qualitative methods .
- If you want to analyze a large amount of readily-available data, use secondary data. If you want data specific to your purposes with control over how it is generated, collect primary data.
- If you want to establish cause-and-effect relationships between variables , use experimental methods. If you want to understand the characteristics of a research subject, use descriptive methods.
Quantitative research deals with numbers and statistics, while qualitative research deals with words and meanings.
Quantitative methods allow you to systematically measure variables and test hypotheses . Qualitative methods allow you to explore concepts and experiences in more detail.
Sources in this article
We strongly encourage students to use sources in their work. You can cite our article (APA Style) or take a deep dive into the articles below.
George, T. (2024, January 12). What is Secondary Research? | Definition, Types, & Examples. Scribbr. Retrieved June 18, 2024, from https://www.scribbr.com/methodology/secondary-research/
Largan, C., & Morris, T. M. (2019). Qualitative Secondary Research: A Step-By-Step Guide (1st ed.). SAGE Publications Ltd.
Peloquin, D., DiMaio, M., Bierer, B., & Barnes, M. (2020). Disruptive and avoidable: GDPR challenges to secondary research uses of data. European Journal of Human Genetics , 28 (6), 697–705. https://doi.org/10.1038/s41431-020-0596-x
Is this article helpful?
Tegan George
Other students also liked, primary research | definition, types, & examples, how to write a literature review | guide, examples, & templates, what is a case study | definition, examples & methods, what is your plagiarism score.
Information
- Author Services
Initiatives
You are accessing a machine-readable page. In order to be human-readable, please install an RSS reader.
All articles published by MDPI are made immediately available worldwide under an open access license. No special permission is required to reuse all or part of the article published by MDPI, including figures and tables. For articles published under an open access Creative Common CC BY license, any part of the article may be reused without permission provided that the original article is clearly cited. For more information, please refer to https://www.mdpi.com/openaccess .
Feature papers represent the most advanced research with significant potential for high impact in the field. A Feature Paper should be a substantial original Article that involves several techniques or approaches, provides an outlook for future research directions and describes possible research applications.
Feature papers are submitted upon individual invitation or recommendation by the scientific editors and must receive positive feedback from the reviewers.
Editor’s Choice articles are based on recommendations by the scientific editors of MDPI journals from around the world. Editors select a small number of articles recently published in the journal that they believe will be particularly interesting to readers, or important in the respective research area. The aim is to provide a snapshot of some of the most exciting work published in the various research areas of the journal.
Original Submission Date Received: .
- Active Journals
- Find a Journal
- Proceedings Series
- For Authors
- For Reviewers
- For Editors
- For Librarians
- For Publishers
- For Societies
- For Conference Organizers
- Open Access Policy
- Institutional Open Access Program
- Special Issues Guidelines
- Editorial Process
- Research and Publication Ethics
- Article Processing Charges
- Testimonials
- Preprints.org
- SciProfiles
- Encyclopedia

Article Menu

- Subscribe SciFeed
- Recommended Articles
- Author Biographies
- Google Scholar
- on Google Scholar
- Table of Contents
Find support for a specific problem in the support section of our website.
Please let us know what you think of our products and services.
Visit our dedicated information section to learn more about MDPI.
JSmol Viewer
Research on evaluation of the carbon dioxide sequestration potential in saline aquifers in the qiongdongnan–yinggehai basin, 1. introduction, 2. evaluation method for carbon sequestration potential in saline aquifers, 2.1. division of evaluation levels at present, 2.2. calculation methods, 3. calculation model and parameter selection, 3.1. sequestration potential calculation model, 3.2. parameter acquisition and processing methods, 3.3. calculation of the sequestration potential, 4. saline aquifer carbon sequestration suitability evaluation, 4.1. necessary indicators, 4.2. key indicators, 5. results of carbon sequestration potential evaluation in saline aquifers, 5.1. carbon sequestration potential and suitability evaluation of the yinggehai basin, 5.2. carbon sequestration potential and suitability evaluation of the qiongdongnan basin, 6. conclusions, author contributions, data availability statement, conflicts of interest.
- Baker, E.; Chon, H.; Keisler, J. Carbon capture and storage: Combining economic analysis with expert elicitations to inform climate policy. Clim. Chang. 2009 , 96 , 379–408. [ Google Scholar ] [ CrossRef ]
- Haszeldine, R.S. Carbon capture and storage: How green can black be? Science 2009 , 325 , 1647–1652. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Luo, J.; Xie, Y.; Hou, M.Z.; Xiong, Y.; Wu, X.; Lüddeke, C.; Huang, L. Advances in subsea carbon dioxide utilization and storage. Energy Rev. 2023 , 2 , 100016. [ Google Scholar ] [ CrossRef ]
- Bashir, A.; Ali, M.; Patil, S.; Aljawad, M.S.; Mahmoud, M.; Al-Shehri, D.; Hoteit, H.; Kamal, M.S. Comprehensive review of CO 2 geological storage: Exploring principles, mechanisms, and prospects. Earth-Sci. Rev. 2024 , 249 , 104672. [ Google Scholar ] [ CrossRef ]
- Matter, J.; Kelemen, P. Permanent storage of carbon dioxide in geological reservoirs by mineral carbonation. Nat. Geosci 2009 , 2 , 837–841. [ Google Scholar ] [ CrossRef ]
- Bauer, S.; Class, H.; Ebert, M.; Gotize, H.; Holzheid, A.; Kolditz, O.; Rosenbaum, S.; Rabel, W.; Schafer, D.; Dahmke, A. Modeling, parameterization and evaluation of monitoring methods for CO 2 storage in deep saline formations: The CO 2 -MoPa project. Environ. Earth Sci. 2012 , 67 , 351–367. [ Google Scholar ] [ CrossRef ]
- Bachu, S. Review of CO 2 storage efficiency in deep saline aquifers. Int. J. Greenh. Gas Control 2015 , 40 , 188–202. [ Google Scholar ] [ CrossRef ]
- Kovscek, A.R.; Wang, Y. Geologic storage of carbon dioxide and enhanced oil recovery. i. uncertainty quantification employing a streamline based proxy for reservoir flow simulation. Energy Convers. Manag. 2005 , 46 , 1920–1940. [ Google Scholar ] [ CrossRef ]
- Adu, E.; Zhang, Y.; Liu, D. Current situation of carbon dioxide capture, storage, and enhanced oil recovery in the oil and gas industry. Can. J. Chem. Eng. 2018 , 97 , 1048–1076. [ Google Scholar ] [ CrossRef ]
- Ning, W.; Zunsheng, J.; Kevin, E.; Ku, A.Y.; Shengnan, L.; Richard, M.; Li, X. Decarbonizing the coal-fired power sector in china via carbon capture, geological utilization, and storage technology. Environ. Sci. Technol. 2021 , 55 , 13164–13173. [ Google Scholar ] [ CrossRef ]
- Zhang, L.; Song, Y.; Shi, J.; Shen, Q.; Deng, H.U.; Gao, Q.; Chen, W.; Kow, K.; Pang, C.; Sun, N.; et al. Frontiers of CO 2 capture and utilization (ccu) towards carbon neutrality. Adv. Atmos. Sci. 2022 , 39 , 1252–1270. [ Google Scholar ] [ CrossRef ]
- Global CCS Institute. Globa Status of CCS 2023—eport & Executive Summary. 2023. Available online: https://www.globalccsinstitute.com/resources/publications-reports-research/global-status-of-ccs-2023-executive-summary/ (accessed on 10 June 2024).
- Zhang, L.; Li, D.X.; Ezekiel, J.; Zhang, W.D.; Mi, H.G.; Ren, S.R. CO 2 geological storage into a lateral aquifer of an offshore gas field in the south china sea: Storage safety and project design. Front. Earth Sci. 2015 , 9 , 286–299. [ Google Scholar ] [ CrossRef ]
- Baines, S.J.; Worden, R.H. Geological storage of carbon dioxide. Rud.-Geološko-Naft. Zb. 2004 , 28 , 9–22. [ Google Scholar ] [ CrossRef ]
- Leung, D.Y.C.; Caramanna, G.; Maroto-Valer, M.M. An overview of current status of carbon dioxide capture and storage technologies. Renew. Sustain. Energy Rev. 2014 , 39 , 426–443. [ Google Scholar ] [ CrossRef ]
- Li, L.; Zhao, N.; Wei, W.; Sun, Y. A review of research progress on CO 2 capture, storage, and utilization in chinese academy of sciences. Fuel 2013 , 108 , 112–130. [ Google Scholar ] [ CrossRef ]
- Zhang, L.; Ezekiel, J.; Li, D.; Pei, J.; Ren, S. Potential assessment of CO 2 injection for heat mining and geological storage in geothermal reservoirs of china. Appl. Energy 2014 , 122 , 237–246. [ Google Scholar ] [ CrossRef ]
- Gibbins, J.; Chalmers, H. Carbon capture and storage. energy policy. Energy Policy 2008 , 36 , 4317–4322. [ Google Scholar ] [ CrossRef ]
- Zhang, X.; Fan, J.L.; Wei, Y.M. Technology roadmap study on carbon capture, utilization and storage in China. Energy Policy 2013 , 59 , 536–550. [ Google Scholar ] [ CrossRef ]
- Goodman, A.; Bromhal, G.; Strazisar, B.; Rodosta, T.; Guthrie, W.F.; Allend, D. Comparison of methods for geologic storage of carbon dioxide in saline formations. Int. J. Greenh. Gas Control 2013 , 18 , 329–342. [ Google Scholar ] [ CrossRef ]
- Diao, Y.J.; Liu, T.; Wei, N.; Ma, X.; Jin, X.L.; Fu, L. Classification and assessment methodology of carbon dioxide geological storage in deep saline aquifers. Geol. China 2023 , 50 , 943–951. [ Google Scholar ] [ CrossRef ]
- Szulczewski, M.L.; MacMinn, C.W.; Herzog, H.J.; Juanes, R. Lifetime of carbon capture and storage as a climate-change mitigation technology. Proc. Natl. Acad. Sci. USA 2012 , 109 , 5185–5189. [ Google Scholar ] [ CrossRef ]
- Zhou, Q.; Birkholzer, J.T.; Tsang, C.-F.; Rutqvist, J. A method for quick assessment of CO 2 storage capacity in closed and semi-closed saline formation. Greenh. Gas Control 2008 , 2 , 626–639. [ Google Scholar ] [ CrossRef ]
- Shen, P.; Liao, X.; Liu, Q. Methodology for estimation of CO 2 storage capacity in reservoirs. Pet. Explor. Dev. 2009 , 36 , 216–220. [ Google Scholar ] [ CrossRef ]
- Brennan, S.T.; Burruss, R.C.; Merrill, M.D.; Freeman, P.A.; Ruppert, L.F. A Probabilistic Assessment Methodology for the Evaluation of Geologic Carbon Dioxide Storage ; Open File Report 1127; U. S. Geological Survey: Sunrise Valley Drive Reston, VA, USA, 2010; pp. 1–39. Available online: https://www.researchgate.net/publication/262802350_A_Probabilistic_Assessment_Methodology_for_the_Evaluation_of_Geologic_Carbon_Dioxide_Storage (accessed on 10 June 2024).
- US-DOE-NETL. Carbon Sequestration Atlas of the United State and Canada , 2nd ed.; U.S. Deparment of Energy—National Energy Technology Laboratory—Office of Fossil Energy: Albany, OR, USA, 2008. Available online: http://www.netl.doe.gov/technologies/carbonseq/refshelf/atlas/ (accessed on 10 June 2024).
- IEA-GHG. Development of Storage Coefficients for CO2 Storage in Deep Saline Formations ; IEA Greeen house Gas R&D Programme (IEA-GHG), Report Number 2009/13; IEA-GHG: Cheltenham, UK, 2009; Available online: https://www.globalccsinstitute.com/resources/publications-reports-research/development-of-storage-coefficients-for-carbon-dioxide-storage-in-deep-saline-formations/ (accessed on 10 June 2024).
- Gorecki, C.D.; Sorensen, J.A.; Bremer, J.M.; Ayash, S.C.; Harju, J.A. Development of Storage Coefficients for Carbon Dioxide Storage in Deep Saline Formations. In Proceedings of the SPE International Conference on CO2 Capture, Storage, and Utilization, San Diego, CA, USA, 2–4 November 2009. [ Google Scholar ] [ CrossRef ]
- Goodman, A.; Hakala, A.; Bromhal, G.; Deel, D.; Rodosta, T.; Frailey, S.; Small, M.; Allen, D.; Romanov, V.; Fazio, J.; et al. U.S. DOE methodology for the development of geologic storage potential for carbon dioxide at the national and regional scale. Int. J. Greenh. Gas Control 2011 , 5 , 952–965. [ Google Scholar ] [ CrossRef ]
- Anwar, S.; Carroll, J.J. Carbon Dioxide Thermodynamic Properties Handbook: Covering Temperatures from −20 ° to 250 °C and Pressures up to 1000 Bar , 2nd ed.; Wiley: West Maitland, FL, USA, 2016. [ Google Scholar ] [ CrossRef ]
- Cai, B.; Li, Q.; Liu, G.; Liu, L.; Jin, T.; Shi, H. Environmental concern-based site screening of carbon dioxide geological storage in China. Sci. Rep. 2017 , 7 , 7598. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Mia, Z.; Wang, F.; Yang, Y.; Wang, F.; Hua, T.; Tian, H.; Tian, A. Evaluation of the potentiality and suitability for CO 2 geological storage in the Junggar Basin, northwestern China. Int. J. Greenh. Gas Control 2018 , 78 , 62–72. [ Google Scholar ] [ CrossRef ]
- Bachu, S. Screening and ranking of sedimentary basins for sequestration of CO 2 in geological media in response to climate change. Environ. Geol. 2003 , 44 , 277–289. [ Google Scholar ] [ CrossRef ]
- Anthonsen, K.L.; Aagaard, P.; Bergmo, P.E.S.; Erlström, M.; Fareide, J.I.; Gislason, S.R.; Mortensen, G.M.; Snæbjörnsdottir, S.Ó. CO 2 storage potential in the Nordic region. Energy Procedia 2013 , 37 , 5080–5092. [ Google Scholar ] [ CrossRef ]
- Vangkilde-Pedersen, A.; Anghe, A.; Zivkovic, A. Wp2 Report—Storage Capacity. eu Geocapacity Project—Assessing European Capacity for Geological Storage of Carbon Dioxide. 2009. Available online: https://www.researchgate.net/publication/262688927_WP2_Report_-_Storage_capacity_EU_GeoCapacity_project_-_Assessing_European_Capacity_for_Geological_Storage_of_Carbon_Dioxide (accessed on 10 June 2024).
- Oldenburg, C.M. Health, Safety, and Environmental Screening and Ranking Framework for Geologic CO 2 Storage Site Selection ; Lawrence Berkeley National Laboratory: Berkeley, CA, USA, 2005. [ Google Scholar ] [ CrossRef ]
- Huo, C.L. Study on the Potential Evaluation and the Storage Areas of the Carbon Dioxide Seabed Storage in Offshore China. Ph.D. Dissertation, Dalian Maritime University, Dalian, China, 2014. [ Google Scholar ]
- He, J.; Zhang, W.; Lu, Z. Seepage system of oil-gas and its exploration in Yinggehai Basin located at northwest of South China Sea. J. Nat. Gas Geosci. 2017 , 2 , 29–41. [ Google Scholar ] [ CrossRef ]
- Zhu, Y.; Sun, L.; Hao, F.; Tu, L. Geochemical composition and origin of Tertiary oils in the Yinggehai and Qiongdongnan Basins, offshore South China Sea. Mar. Pet. Geol. 2018 , 96 , 139–153. [ Google Scholar ] [ CrossRef ]
- Zhao, J.; Li, J.; Xu, Z. Advances in the origin of overpressures in sedimentary basins. Pet. Res. 2018 , 3 , 1–24. [ Google Scholar ] [ CrossRef ]
- Zhang, G.C.; Zhang, Y.; Shen, H.; He, Y. An analysis of natural gas exploration potential in the Qiongdongnan Basin by use of the theory of “joint control of source rocks and geothermal heat”. Nat. Gas Ind. B 2014 , 1 , 41–50. [ Google Scholar ] [ CrossRef ]
- Yu, J.; Pei, J.; Xu, J. New insight into oil and gas exploration in Miocene and Late Oligocene strata in Qiongdongnan basin. J. Earth Sci. 2009 , 20 , 811–823. [ Google Scholar ] [ CrossRef ]
Click here to enlarge figure
| Evaluation Level | Applicable Conditions | Estimation Method |
|---|---|---|
| E | Basin level | Mechanism Method (Forum Geological Work Group) [ ] Volumetric Method (U.S. Department of Energy Geological Task Force; U.S. Geological Survey) [ ] |
| D | Zone level | Mechanism Method (Forum Geological Work Group) Volumetric Method (U.S. Department of Energy Geological Task Force; U.S. Geological Survey) |
| C | Target district level | Mechanism Method (Forum Geological Work Group) Volumetric Method (U.S. Department of Energy Geological Task Force; U.S. Geological Survey) |
| B | Site level | Mechanism Method (Forum Geological Work Group) [ , ] |
| A | Perfusion level | Mechanism Method |
| Indicator Level | First-Level Indicators (Basin Weight/Zone Weight) | Second-Level Indicators (Basin Indicator Weight) | Second-Level Indicators (Zone Indicator Weight) | Description |
|---|---|---|---|---|
| Necessary Indicators | Sequestration Potential (0.3/0.4) | Basin Area (0.05) | Zone Area (0.1) | The area of the basin/zone projected onto the plane. |
| Basin Thickness (0.05) | Zone Thickness (0.1) | The thickness of Cenozoic strata buried between 800 and 3200 m in the basin/zone. The thicker the strata, the more favorable it is for CO geological sequestration. The burial depth also affects the implementation conditions. | ||
| Sequestration Potential (0.1) | Sequestration Potential (0.1) | The predicted or presumed potential of the basin/zone for CO geological sequestration. The greater the sequestration potential, the more suitable it is for CO geological sequestration. | ||
| Per Unit Area Sequestration Potential (0.1) | Per Unit Area Sequestration Potential (0.1) | The potential amount of CO that can be sequestered per unit area in the basin/zone. | ||
| Geological Conditions(0.3/0.3) | Exploration Degree (0.1) | Exploration Degree (0.05) | This reflects the level of knowledge and data richness of the sedimentary basin/zone. The higher the degree of exploration, the more reliable and accurate the evaluation indicators become. This is beneficial for accurately assessing the suitability of CO geological sequestration in the sedimentary basin/zone. | |
| Seafloor Temperature (0.05) | Seafloor Temperature (0.05) | The average temperature of seawater at the seabed. The temperature of seawater has a certain impact on geothermal energy. For a marine region, the temperature of the seafloor primarily depends on the latitude and water depth, and the average value over multiple years is considered. | ||
| Geothermal Gradient (0.05) | Geothermal Gradient (0.05) | Expressed as the number of degrees Celsius (°C) of temperature increase per 100 m of vertical depth. This indicator reflects the rate of temperature increase within the strata with depth and is one of the important parameters that affect the potential of CO geological sequestration. The geothermal gradient is determined by the Earth’s internal heat and the thermal conductivity of the strata. | ||
| Fault activity (0.1) | Fault Activity (0.05) | Divided into three categories: inactive faults, faults without through-going faults, and faults with through-going faults. | ||
| Reservoir Conditions (0.05) | Based on the size of porosity, the reservoir carbon layers are divided into three categories: high-quality, good, and effective carbon storage layers. | |||
| Cap Rock Conditions (0.05) | Cap rocks are classified based on their thickness and scale. | |||
| Engineering Conditions (0.4/0.3) | Development Degree (0.1) | Development Degree (0.1) | This reflects the extent of oil and gas development in the basin/zone. The higher the level of development, the more advanced the drilling platforms and pipeline network become, leading to better engineering conditions. | |
| Offshore Distance (0.1) | Offshore Distance (0.1) | The shortest distance from the basin/zone to the coast. The farther the distance, the higher the transportation and injection costs, the greater the technical difficulty, and the less favorable it is for CO geological sequestration. | ||
| Seawater Depth (0.2) | Seawater Depth (0.1) | When the depth of seawater exceeds 150 m, more sophisticated and costly platforms and processes are necessary. The greater the depth, the higher the technical difficulty, and the greater the cost. | ||
| Key Indicators | Seismic Belt | Based on the occurrence of earthquakes above a magnitude of 8 in a basin or zone over the past century, it is divided into seismic belts. If the condition applies, a value of 0 is assigned; otherwise, a value of 1 is assigned. | ||
| Drilling Engineering Feasibility | The feasibility of drilling engineering is primarily determined by whether oil and gas drilling projects have been implemented in the basin or zone. If there is no oil and gas drilling, this indicator is assigned a value of 0; otherwise, it is assigned a value of 1. | |||
| Basin and Zone | Geothermal Gradient (°C/km) | Seafloor Temperature (°C) | Water Depth (m) |
|---|---|---|---|
| Yinggehai Basin | 40 | 24 | 50 |
| Central Depression | 36 | 24 | 50 |
| Eastern Slope | 39 | 24 | 50 |
| Western Slope | 39 | 24 | 50 |
| Basin and Zone | Geothermal Gradient (°C/km) | Seafloor Temperature (°C) | Water Depth (km) |
|---|---|---|---|
| Qiongdongnan Basin | 40 | 5 | 1 |
| Central Uplift | 39 | 15 | 0.15 |
| Central Depression | 40 | 5 | 1 |
| Northern Depression | 37 | 20 | 0.1 |
| Level | Basin and Zone | Area (within National Boundaries) km | Porosity | Sand-to-Shale Ratio | CO Density (kg/m ) | Effective Sequestration Potential of Basin/Zone (×10 t) | Sequestration Potential per Unit Area (×10 t/km ) | ||
|---|---|---|---|---|---|---|---|---|---|
| E1 = 1.2% | E2 = 2.4% | E3 = 4.1% | |||||||
| Basin Level | Yinggehai Basin | 46,929 | 0.11–0.29 | 0.3 | 263–518 | 303 | 606 | 1036 | 129 |
| Zone Level | Central Depression | 35,273 | 0.31 | 289–560 | 273 | 546 | 932 | 155 | |
| Eastern Slope | 10,437 | 0.29 | 269–528 | 66 | 131 | 224 | 126 | ||
| Western Slope | 1218 | 0.29 | 269–528 | 7 | 13 | 22 | 108 | ||
| Basin Level | Qiongdongnan Basin | 96,289 | 0.18–0.26 | 0.32 | 678–822 | 1307 | 2615 | 4467 | 272 |
| Zone Level | Central Uplift | 13,119 | 0.34 | 415–573 | 133 | 265 | 453 | 202 | |
| Central Depression | 71,466 | 0.3 | 678–822 | 972 | 1943 | 3320 | 272 | ||
| Northern Depression | 11,704 | 0.34 | 353–568 | 70 | 140 | 240 | 120 | ||
| The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
Share and Cite
Tian, Y.; Du, Z.; Zhang, L.; Zhang, L.; Xu, G.; Chen, J. Research on Evaluation of the Carbon Dioxide Sequestration Potential in Saline Aquifers in the Qiongdongnan–Yinggehai Basin. J. Mar. Sci. Eng. 2024 , 12 , 997. https://doi.org/10.3390/jmse12060997
Tian Y, Du Z, Zhang L, Zhang L, Xu G, Chen J. Research on Evaluation of the Carbon Dioxide Sequestration Potential in Saline Aquifers in the Qiongdongnan–Yinggehai Basin. Journal of Marine Science and Engineering . 2024; 12(6):997. https://doi.org/10.3390/jmse12060997
Tian, Yukun, Zhili Du, Lin Zhang, Lizhong Zhang, Guoqiang Xu, and Jiaojiao Chen. 2024. "Research on Evaluation of the Carbon Dioxide Sequestration Potential in Saline Aquifers in the Qiongdongnan–Yinggehai Basin" Journal of Marine Science and Engineering 12, no. 6: 997. https://doi.org/10.3390/jmse12060997
Article Metrics
Article access statistics, further information, mdpi initiatives, follow mdpi.

Subscribe to receive issue release notifications and newsletters from MDPI journals

IMAGES
VIDEO
COMMENTS
Learn about primary, secondary, tertiary, and emerging data sources for research, with definitions and examples. Find out how to collect and analyze data effectively for your study objectives.
Learn about the definition, structure, types, and examples of research methodology for different research purposes and designs. Find out how to collect, analyze, and interpret data using quantitative, qualitative, or mixed methods.
Learn how to collect data for your research project using different methods, such as surveys, interviews, experiments, observations, and secondary data. Find out how to plan, operationalize, sample, and standardize your data collection procedures.
Learn how to choose and use different methods for collecting and analyzing data in your research. Compare qualitative and quantitative, primary and secondary, descriptive and experimental data collection methods.
Learn about the different types of research data, such as quantitative, qualitative, primary and secondary data, and their formats, such as text, numeric, audio, video, etc. Explore the common methods of data collection and analysis, such as surveys, interviews, experiments, descriptive statistics, inferential statistics, content analysis, etc.
Learn how to write a research methodology chapter for your thesis, dissertation, or research paper. Find out what to include, how to explain your data collection and analysis methods, and why they are important.
This chapter discusses the methodology and sources of data for a mixed method research on disaster, gender and vulnerability in the coastal area of Bangladesh. It explains the rationale, design, methods, challenges and limitations of the study, and provides a map of the research settings.
A data source is any location where you can find facts, figures, or other relevant information to support your research. You may create your own data source through experimentation, surveys, or observations, or you may choose to use data produced by other researchers.Both methods have advantages and disadvantages, depending on your research and the quality of the existing data you can find.
We set a clear distinction between overall research methodology and the data analysis method. The qualitative analysis method is only a small part of the entire qualitative research methodology. ... At first, data "dumping" from various websites and data sources to local drive is needed to obtain a collection of data, regardless of their ...
This chapter presents the quantitative and qualitative methods used to evaluate the development and implementation of a near real-time survey for improving relational aspects of care. The chapter provides an overview of the data sources and methods used to answer the four RQs guiding this work. Additional details are presented for each data collection activity, including sampling and ...
SOURCES OF DA TA A ND THEIR EVA LUA TION METHODOLOGY. 14.1 INTRODUCTION. The task of data collection begins after a research problem has been defined and. research design/ plan chalked out. While ...
The purpose of primary data is to gather information directly from the source, without relying on secondary sources or pre-existing data. This data is collected through research methods such as surveys, interviews, experiments, and observations. Primary data is valuable because it is tailored to the specific research question or problem at hand ...
Learn how to choose and use research methods for collecting and analysing data. Compare qualitative and quantitative, primary and secondary, descriptive and experimental methods with examples and pros and cons.
The pre-defined topics in the interview guide can be derived from the literature, previous research or a preliminary method of data collection, e.g. document study or observations. The topic list is usually adapted and improved at the start of the data collection process as the interviewer learns more about the field [ 20 ].
This chapter discusses the methodology of a mixed method research design to study disaster, gender and vulnerability in the coastal area of Bangladesh. It explains the rationale, data collection, analysis and limitations of the study, and provides a map of the research settings.
About Research Methods. This guide provides an overview of research methods, how to choose and use them, and supports and resources at UC Berkeley. As Patten and Newhart note in the book Understanding Research Methods, "Research methods are the building blocks of the scientific enterprise. They are the "how" for building systematic knowledge.
Qualitative research methods. Each of the research approaches involve using one or more data collection methods.These are some of the most common qualitative methods: Observations: recording what you have seen, heard, or encountered in detailed field notes. Interviews: personally asking people questions in one-on-one conversations. Focus groups: asking questions and generating discussion among ...
There are two main groups of research methods in the social sciences: ... If other data sources exist, explain why the data you chose is most appropriate to addressing the research problem. Provide a justification for case study selection. A common method of analyzing research problems in the social sciences is to analyze specific cases.
Mixed-method approaches combine both qualitative and quantitative methods, and therefore combine the strengths of both types of research. Mixed methods have gained popularity in recent years. When undertaking mixed-methods research you can collect the qualitative and quantitative data either concurrently or sequentially.
There should be a section on the chosen methodology and a brief discussion about why qualitative methodology was most appropriate for the study question and why one particular methodology (e.g., interpretative phenomenological analysis rather than grounded theory) was selected to guide the research. The method itself should then be described ...
Step 2: Choose your data collection method. Based on the data you want to collect, decide which method is best suited for your research. Experimental research is primarily a quantitative method. Interviews, focus groups, and ethnographies are qualitative methods. Surveys, observations, archival research, and secondary data collection can be ...
Quantitative methods are best for numerical data, while qualitative methods are suitable for textual or thematic data. Understand the Purpose of Each Methodology. Becoming familiar with the four types of research - descriptive, correlational, experimental, and diagnostic - will enable you to select the most appropriate method for your ...
About the Data. Data Sources. Methodology. ... The independent source for health policy research, polling, and news, ...
Collect the raw data sets you'll need to help you answer the identified question. Data collection might come from internal sources, like a company's client relationship management (CRM) software, or from secondary sources, like government records or social media application programming interfaces (APIs). Clean the data to prepare it for ...
Primary Research | Definition, Types, & Examples. Published on January 14, 2023 by Tegan George.Revised on January 12, 2024. Primary research is a research method that relies on direct data collection, rather than relying on data that's already been collected by someone else.In other words, primary research is any type of research that you undertake yourself, firsthand, while using data that ...
What is a Research Design? A research design is defined as the overall plan or structure that guides the process of conducting research. It is a critical component of the research process and serves as a blueprint for how a study will be carried out, including the methods and techniques that will be used to collect and analyze data.
Source Data. Full size image. ... (Methods and Extended Data Fig. 3), we synthesized a pool of overlapping oligonucleotides to tile the 2 kb region containing all candidate SNPs, and included ...
Secondary research is a research method that uses data that was collected by someone else. In other words, whenever you conduct research using data that already exists, you are conducting secondary research. On the other hand, any type of research that you undertake yourself is called primary research. Example: Secondary research.
This paper evaluates the carbon dioxide sequestration potential in the saline aquifers of the South Qiongdongnan-Yinggehai Basin. By using a hierarchical evaluation method, the assessment is divided into five stages: the basin level, the zone level, the target level, the site level, and the injection level. The study primarily focuses on evaluating the sequestration potential of and ...